
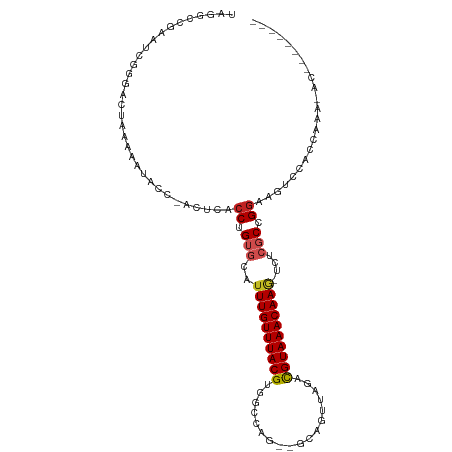
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 5,637,788 – 5,637,892 |
| Length | 104 |
| Max. P | 0.598440 |

| Location | 5,637,788 – 5,637,892 |
|---|---|
| Length | 104 |
| Sequences | 11 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 69.31 |
| Shannon entropy | 0.63927 |
| G+C content | 0.48008 |
| Mean single sequence MFE | -26.73 |
| Consensus MFE | -11.13 |
| Energy contribution | -11.45 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.10 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.598440 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 5637788 104 - 21146708 UAGGCCGAAUCGGUCCUAAAAAUACC-ACUCACCUGUGCAUUUGUUUACGUGGCUGG--GCCGUUAGACGUAAACAAG-UCUCGCCGGAAGUCCACGAAAGACAGAAA--- (((((((...))).)))).......(-((......)))..(((((((.(((((((..--.((((.((((........)-))).).))).)).)))))..)))))))..--- ( -30.20, z-score = -1.37, R) >droEre2.scaffold_4929 17914690 98 + 26641161 UAGGCCGAAUCGAACCUAAAAAUAUC-ACUCACCUGUGCAUUUGUUUACGUGGCC-G--GCCGUUGGGUGUAAACAAG-UCUCGCCGGAAGUCCACCAAAGAC-------- ..(((((......((.((((...(((-((......))).))...)))).))...)-)--))).(((((((........-...))))((....))..)))....-------- ( -20.10, z-score = 1.20, R) >droYak2.chr2L 18263729 105 - 22324452 UAGGCCGAAUCGGGCCUAAAAAUACC-GCUCACCUGUGCAUUUGUUUACGUGGCCGG--G-CGUUAGAUGUAAACAAG-UCUCGCCGGAAGUCCACGAAGUCCUCAAGAC- ((((((......))))))........-((.(....).)).........(((((((((--.-((..((((........)-)))))))))....)))))..(((.....)))- ( -28.80, z-score = -0.15, R) >droSec1.super_1 3247007 104 - 14215200 UAGGCCGAAUCGGGACUAAAAAUAUC-ACUCACCUGUGCAUUUGUUUACGUGGCCGG--GCCGUUAGACGUAAACAAG-UCUCGCCGGAAGUCCACGAAAGACAGAAA--- ....(((...(((((((........(-((......)))....((((((((((((...--))).....)))))))))))-))))).)))..(((.......))).....--- ( -27.10, z-score = -0.30, R) >droSim1.chr2R 4273055 104 - 19596830 UAGGCCGAAUCGGGUCUAAAAAUACC-ACUCACCUGUGCAUUUGUUUACGUGGCCGG--GCCGUUAGACGUAAACAAG-UCUCGCCGGAAGUCCACGAAAGACAGAAA--- ((((((......)))))).......(-((......)))..(((((((.(((((.(..--.((((.((((........)-))).).)))..).)))))..)))))))..--- ( -29.80, z-score = -0.90, R) >droAna3.scaffold_13266 12909066 99 - 19884421 UGGGCCGUGUUCUGGCCAAAAAUACG-ACUCACCUGUGCAUUUGUUUACGCGACCAG--CUAGUUAGACGUAAACAAG-UCUCGCCGGAAGUGGGCCAGCAAC-------- ..((((...(((((((......((((-.......)))).(((((((((((((((...--...))).).))))))))))-)...)))))))...))))......-------- ( -29.60, z-score = -0.49, R) >droMoj3.scaffold_6496 2767859 98 + 26866924 -----------CAAAGCACCUAUAAAUACUUACCUGAACAUUUGUUUACGUACCCAG-CCCAAUUAGACGUAAACAAG-AGCCGCCGGAAGUAGGCCACGUAAAAAAAAAA -----------..................((((.((....(((((((((((......-.........)))))))))))-.(((..(....)..))))).))))........ ( -19.66, z-score = -2.51, R) >droGri2.scaffold_15112 871443 78 - 5172618 -----------UCGAGCACCUAUAAAUACUCAGCUGACCAUUUGUUUACGUGGGCAG--CCAAUUAGACGUAAACAAC-AGUCGCCGGAAGU------------------- -----------(((.((...............((((.((((........)))).)))--)......(((.........-.))))))))....------------------- ( -16.20, z-score = -0.44, R) >droWil1.scaffold_180701 3605295 92 - 3904529 -------GGUUUAGUUUAUAAAUAAA-AUUCACCUGUGCAUUUGUUUACGUGGGCCGUACCAAUUAGACGUAAACAAAUCAACGCCGGAAGUGGGCCAAA----------- -------((((((.((((....))))-.....((.(((.((((((((((((((......))......))))))))))))...))).))...))))))...----------- ( -21.20, z-score = -0.47, R) >droPer1.super_4 6108479 96 + 7162766 UUGGCCAGAGUCUGGGUAUAAAUAGG-ACUCACCUGUGCAUUUGUUUACGCGACCAG--UCAGUUAGACGUAAACAAG-UCUCGCCGGAAGUGGGCCAAA----------- ((((((.((((((..((....)).))-)))).((.(((.(((((((((((((((...--...))).).))))))))))-)..))).)).....)))))).----------- ( -34.50, z-score = -2.92, R) >dp4.chr3 14261643 96 - 19779522 UUGGCCAGAGUCUGGCUAUAAAUAGG-ACUCACCUGUGCAUUUGUUUACGCGACCAG--UCAGUUAGACGUAAACAAG-UCUCGCCGGAAGUGGGCCAAA----------- ((((((....((((((.....(((((-.....)))))..(((((((((((((((...--...))).).))))))))))-)...))))))....)))))).----------- ( -36.90, z-score = -3.73, R) >consensus UAGGCCGAAUCGGGACUAAAAAUACC_ACUCACCUGUGCAUUUGUUUACGUGGCCAG__GCAGUUAGACGUAAACAAG_UCUCGCCGGAAGUCCACCAAA_AC________ ................................((.(((..((((((((((..................))))))))))....))).))....................... (-11.13 = -11.45 + 0.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:10:53 2011