
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 5,613,253 – 5,613,351 |
| Length | 98 |
| Max. P | 0.960796 |

| Location | 5,613,253 – 5,613,351 |
|---|---|
| Length | 98 |
| Sequences | 9 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 78.08 |
| Shannon entropy | 0.43367 |
| G+C content | 0.44936 |
| Mean single sequence MFE | -35.08 |
| Consensus MFE | -16.78 |
| Energy contribution | -16.95 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.45 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.940364 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 5613253 98 + 21146708 ----------AGUGAGCGAGUGAGUGCAUGUGUGAGUGCAUUU-GAGUGCACUCUGCCAAUAAUGCGCACAAUUUAAGUUUCGGCAAACACGCCCAAACACUCGCAUUA ----------.....((((((..((((((.((.((((((((..-..))))))))...))...)))))).........((((.(((......))).)))))))))).... ( -39.20, z-score = -3.82, R) >droSim1.chr2R 4254951 98 + 19596830 ----------AGUGAGCGAGUGAGUGCAUGUGUGAGUGCAUUU-GAGUGCACUCUGCCAAUAAUGCGCACAAUUUAAGUUUCGGCAAACACGCCCAAACACUCGCAUUA ----------.....((((((..((((((.((.((((((((..-..))))))))...))...)))))).........((((.(((......))).)))))))))).... ( -39.20, z-score = -3.82, R) >droSec1.super_1 3230097 98 + 14215200 ----------AGUGAGCGAGUGAGUGCAUGUGUGAGUGCAUUU-GAGUGCACUCUGCCAAUAAUGCGCACAAUUUAAGUUUCGGCAAACACGCCCAAACACUCGCAUUA ----------.....((((((..((((((.((.((((((((..-..))))))))...))...)))))).........((((.(((......))).)))))))))).... ( -39.20, z-score = -3.82, R) >droYak2.chr2L 18246627 94 + 22324452 --------------AGUGAGUGAGUGCAUGUAUGAGUGCAUUU-GAGUGCACUCUGCCAAUAAUGCGCACAAUUUAAGUUUCGGCAAACACGCCCAGGCACUGGCAUUA --------------.((.(((..(((((((((.((((((((..-..))))))))))).....)))))).........((((.(((......))).))))))).)).... ( -32.60, z-score = -1.75, R) >droEre2.scaffold_4929 17898080 94 - 26641161 --------------AGUGAGUGAGUGCAUGUGUGAGUACAUUU-GAGUGCACUCUGCCAAUAAUGCGCACAAUUUAAGUUUCGGCAAACACGUCCAAACAGCCGCAUUA --------------.(((.((((((((((..(((....)))..-..))))))))(((((....)).)))........((((.(((......))).)))).))))).... ( -23.40, z-score = 0.27, R) >droAna3.scaffold_13266 12893794 89 + 19884421 ----------AGUGAGUGUGCGAGUGAAUGUGUGAGUGCAUUUUGAGUGCACUCUGCCAAUAAUGCGCACAAUUUAAGUUUCGGCAAACACUUGCAUUA---------- ----------.(..(((((.(((((((((((((((((((((.....)))))))).((.......)))))).)))))...))))....)))))..)....---------- ( -29.90, z-score = -1.93, R) >dp4.chr3 14244141 108 + 19779522 AGAGCGAGUGUGUGUGCAUGUGCAUGUGCAUGUGAGUGCAUUU-GAGUGCACUCUGCCAAUAAUGCGCACAAUUUAAGUUUCGGAAAACACGCCCAAACACUUGCAUUA ...((((((((((((((((((((....))))..((((((((..-..))))))))........)))))))).......((((....))))........)))))))).... ( -40.80, z-score = -2.67, R) >droPer1.super_4 6090985 108 - 7162766 AGAGCGAGUGUGUGUGCAUGUGCAUGUGCAUGUGAGUGCAUUU-GAGUGCACUCUGCCAAUAAUGCGCACAAUUUAAGUUUCGGCAAACACGCCCAAACACUUGCAUUA ...((((((.(((((((((((((....))))..((((((((..-..))))))))........)))))))))......((((.(((......))).)))))))))).... ( -44.50, z-score = -3.26, R) >droWil1.scaffold_180701 325313 98 - 3904529 -AGUUGUCUGUGUGUGUAUGCGCAUGUGUAUUCGAGUACAUUU-GAGUGCACUCUGCCAAUAAUGCGCACAAUUUAAGUUUCA-CACAAACAUUUGAAUUA-------- -...(((.((((((....((((((((((((((((((....)))-))))))))..........)))))))............))-)))).))).........-------- ( -26.89, z-score = -1.01, R) >consensus __________AGUGAGCGAGUGAGUGCAUGUGUGAGUGCAUUU_GAGUGCACUCUGCCAAUAAUGCGCACAAUUUAAGUUUCGGCAAACACGCCCAAACACUCGCAUUA ...............(((.(((.((........((((((((.....))))))))........)).))).........((((.((........)).))))...))).... (-16.78 = -16.95 + 0.17)

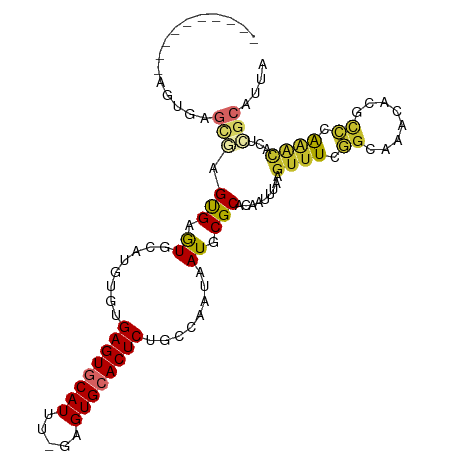
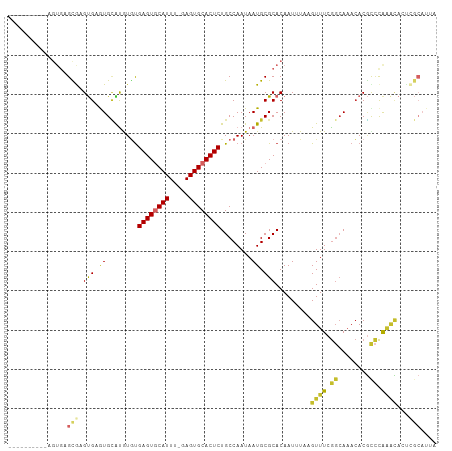
| Location | 5,613,253 – 5,613,351 |
|---|---|
| Length | 98 |
| Sequences | 9 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 78.08 |
| Shannon entropy | 0.43367 |
| G+C content | 0.44936 |
| Mean single sequence MFE | -31.78 |
| Consensus MFE | -16.75 |
| Energy contribution | -16.09 |
| Covariance contribution | -0.65 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.960796 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 5613253 98 - 21146708 UAAUGCGAGUGUUUGGGCGUGUUUGCCGAAACUUAAAUUGUGCGCAUUAUUGGCAGAGUGCACUC-AAAUGCACUCACACAUGCACUCACUCGCUCACU---------- ....((((((((((.((((....)))).)))).......((((((.......)).(((((((...-...)))))))......))))..)))))).....---------- ( -37.20, z-score = -3.61, R) >droSim1.chr2R 4254951 98 - 19596830 UAAUGCGAGUGUUUGGGCGUGUUUGCCGAAACUUAAAUUGUGCGCAUUAUUGGCAGAGUGCACUC-AAAUGCACUCACACAUGCACUCACUCGCUCACU---------- ....((((((((((.((((....)))).)))).......((((((.......)).(((((((...-...)))))))......))))..)))))).....---------- ( -37.20, z-score = -3.61, R) >droSec1.super_1 3230097 98 - 14215200 UAAUGCGAGUGUUUGGGCGUGUUUGCCGAAACUUAAAUUGUGCGCAUUAUUGGCAGAGUGCACUC-AAAUGCACUCACACAUGCACUCACUCGCUCACU---------- ....((((((((((.((((....)))).)))).......((((((.......)).(((((((...-...)))))))......))))..)))))).....---------- ( -37.20, z-score = -3.61, R) >droYak2.chr2L 18246627 94 - 22324452 UAAUGCCAGUGCCUGGGCGUGUUUGCCGAAACUUAAAUUGUGCGCAUUAUUGGCAGAGUGCACUC-AAAUGCACUCAUACAUGCACUCACUCACU-------------- .......((((..(((((((((.((((((....................))))))(((((((...-...)))))))..)))))).).))..))))-------------- ( -27.35, z-score = -0.98, R) >droEre2.scaffold_4929 17898080 94 + 26641161 UAAUGCGGCUGUUUGGACGUGUUUGCCGAAACUUAAAUUGUGCGCAUUAUUGGCAGAGUGCACUC-AAAUGUACUCACACAUGCACUCACUCACU-------------- .....((((...............))))...........((((((.......)).(((((((...-...)))))))......)))).........-------------- ( -21.26, z-score = 0.49, R) >droAna3.scaffold_13266 12893794 89 - 19884421 ----------UAAUGCAAGUGUUUGCCGAAACUUAAAUUGUGCGCAUUAUUGGCAGAGUGCACUCAAAAUGCACUCACACAUUCACUCGCACACUCACU---------- ----------...(((.((((..((...............(((.((....)))))(((((((.......)))))))...))..)))).)))........---------- ( -23.00, z-score = -1.87, R) >dp4.chr3 14244141 108 - 19779522 UAAUGCAAGUGUUUGGGCGUGUUUUCCGAAACUUAAAUUGUGCGCAUUAUUGGCAGAGUGCACUC-AAAUGCACUCACAUGCACAUGCACAUGCACACACACUCGCUCU ....((.(((((.((.((((((................(((((((.......)).(((((((...-...)))))))....)))))...)))))).)).))))).))... ( -34.41, z-score = -1.88, R) >droPer1.super_4 6090985 108 + 7162766 UAAUGCAAGUGUUUGGGCGUGUUUGCCGAAACUUAAAUUGUGCGCAUUAUUGGCAGAGUGCACUC-AAAUGCACUCACAUGCACAUGCACAUGCACACACACUCGCUCU ....((.(((((.((.((((((..(((((........))).))((((.....((((((((((...-...)))))))...)))..)))))))))).)).))))).))... ( -36.00, z-score = -1.70, R) >droWil1.scaffold_180701 325313 98 + 3904529 --------UAAUUCAAAUGUUUGUG-UGAAACUUAAAUUGUGCGCAUUAUUGGCAGAGUGCACUC-AAAUGUACUCGAAUACACAUGCGCAUACACACACAGACAACU- --------.........((((((((-((..........(((((((((........(((((((...-...)))))))........)))))))))..))))))))))...- ( -32.39, z-score = -4.19, R) >consensus UAAUGCGAGUGUUUGGGCGUGUUUGCCGAAACUUAAAUUGUGCGCAUUAUUGGCAGAGUGCACUC_AAAUGCACUCACACAUGCACUCACUCGCUCACU__________ ..................((((.((((((....................))))))(((((((.......)))))))))))............................. (-16.75 = -16.09 + -0.65)
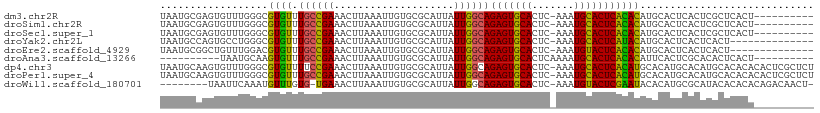
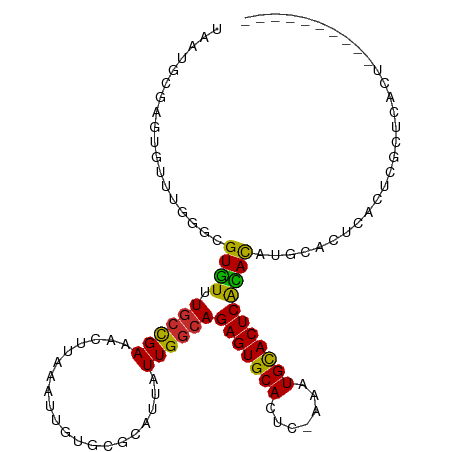
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:10:51 2011