| Sequence ID | dm3.chr2R |
|---|---|
| Location | 5,556,239 – 5,556,329 |
| Length | 90 |
| Max. P | 0.999177 |

| Location | 5,556,239 – 5,556,329 |
|---|---|
| Length | 90 |
| Sequences | 7 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 64.76 |
| Shannon entropy | 0.61062 |
| G+C content | 0.41564 |
| Mean single sequence MFE | -14.15 |
| Consensus MFE | -10.38 |
| Energy contribution | -10.52 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.69 |
| SVM RNA-class probability | 0.999177 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
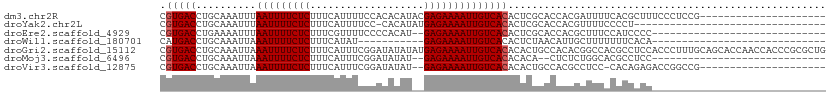
>dm3.chr2R 5556239 90 + 21146708 CGUGACCUGCAAAUUUAAUUUUCUCUUUCAUUUUCCACACAUACGAGAAAAUUGUCACACUCGCACCACGAUUUUCACGCUUUCCCUCCG--------------------- (((((..........((((((((((...................))))))))))......(((.....)))...)))))...........--------------------- ( -12.71, z-score = -2.76, R) >droYak2.chr2L 18187924 78 + 22324452 CGUGACCUGCAAAUUUAAUUUUCUCUUUCAUUUUCC-CACAUAUGAGAAAAUUGUCACACUCGCACCACGUUUUCCCCU-------------------------------- ((((...(((.....((((((((((...........-.......))))))))))........))).)))).........-------------------------------- ( -12.59, z-score = -3.45, R) >droEre2.scaffold_4929 17842233 80 - 26641161 CGUGACCUGAAAAUUUAAUUUUCUCUUUCGUUUUCCCCACAU--GAGAAAAUUGUCACACUCGCACCACGCUUUCCAUCCCC----------------------------- .(((((..........(((((((((.................--))))))))))))))....((.....))...........----------------------------- ( -12.83, z-score = -2.82, R) >droWil1.scaffold_180701 3500210 71 + 3904529 CAUGACCUGCAAAUUAAAUUUUCUCUUUCAUAU-----------GAGAAAAUUGUCACACUCUAACAUUGCUUUUUUUCACA----------------------------- ........((((....(((((((((........-----------)))))))))((.........)).))))...........----------------------------- ( -9.80, z-score = -2.01, R) >droGri2.scaffold_15112 780117 111 + 5172618 CGUGACCUGCAAAUUAAAUUUUCUCUUUCAUUUCGGAUAUAUAUGAGAAAAUUGUCACACACUGCCACACGGCCACGCCUCCACCCUUUGCAGCACCAACCACCCGCGCUG .(((..(((((((...(((((((((...................)))))))))..........(((....))).............))))))))))............... ( -20.01, z-score = -1.57, R) >droMoj3.scaffold_6496 2664759 78 - 26866924 CGUGACCUGCAAAUUAAAUUUUCUCUUUCAUUUCGGAUAUAU--GAGAAAAUUGUCACACACA--CUCUCUGGCACGCCUCC----------------------------- .(((((..........(((((((((.(((.....))).....--)))))))))))))).....--......((....))...----------------------------- ( -13.80, z-score = -1.65, R) >droVir3.scaffold_12875 5855212 87 + 20611582 CGUGACCUGCAAAUUAAAUUUUCUCUUUCAUUUCGGAUAUAU--GAGAAAAUUGUCACACACUGCCACGCCUCC-CACAGAGACCGGCCG--------------------- .(((((..........(((((((((.(((.....))).....--)))))))))))))).....(((..(.(((.-....))).).)))..--------------------- ( -17.30, z-score = -1.60, R) >consensus CGUGACCUGCAAAUUAAAUUUUCUCUUUCAUUUCCCACACAU__GAGAAAAUUGUCACACUCGCACAACGCUUUACACCCCC_____________________________ .(((((..........(((((((((...................))))))))))))))..................................................... (-10.38 = -10.52 + 0.14)
| Location | 5,556,239 – 5,556,329 |
|---|---|
| Length | 90 |
| Sequences | 7 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 64.76 |
| Shannon entropy | 0.61062 |
| G+C content | 0.41564 |
| Mean single sequence MFE | -18.75 |
| Consensus MFE | -10.55 |
| Energy contribution | -10.57 |
| Covariance contribution | 0.02 |
| Combinations/Pair | 1.18 |
| Mean z-score | -0.87 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.824464 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
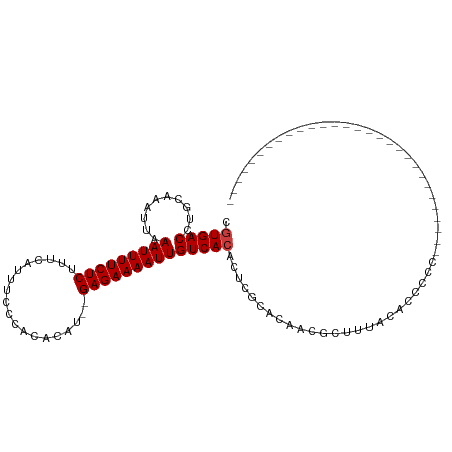
>dm3.chr2R 5556239 90 - 21146708 ---------------------CGGAGGGAAAGCGUGAAAAUCGUGGUGCGAGUGUGACAAUUUUCUCGUAUGUGUGGAAAAUGAAAGAGAAAAUUAAAUUUGCAGGUCACG ---------------------....................((((((((((((.....(((((((((.(((.........)))...)))))))))..)))))))..))))) ( -18.20, z-score = -0.80, R) >droYak2.chr2L 18187924 78 - 22324452 --------------------------------AGGGGAAAACGUGGUGCGAGUGUGACAAUUUUCUCAUAUGUG-GGAAAAUGAAAGAGAAAAUUAAAUUUGCAGGUCACG --------------------------------.........((((((((((((.....(((((((((.(((...-.....)))...)))))))))..)))))))..))))) ( -17.20, z-score = -2.10, R) >droEre2.scaffold_4929 17842233 80 + 26641161 -----------------------------GGGGAUGGAAAGCGUGGUGCGAGUGUGACAAUUUUCUC--AUGUGGGGAAAACGAAAGAGAAAAUUAAAUUUUCAGGUCACG -----------------------------...........((.....))....(((((..(((((((--....))))))).(....).((((((...))))))..))))). ( -16.20, z-score = -0.80, R) >droWil1.scaffold_180701 3500210 71 - 3904529 -----------------------------UGUGAAAAAAAGCAAUGUUAGAGUGUGACAAUUUUCUC-----------AUAUGAAAGAGAAAAUUUAAUUUGCAGGUCAUG -----------------------------.((((......(((((((((.....)))))((((((((-----------........)))))))).....))))...)))). ( -13.20, z-score = -1.41, R) >droGri2.scaffold_15112 780117 111 - 5172618 CAGCGCGGGUGGUUGGUGCUGCAAAGGGUGGAGGCGUGGCCGUGUGGCAGUGUGUGACAAUUUUCUCAUAUAUAUCCGAAAUGAAAGAGAAAAUUUAAUUUGCAGGUCACG ((((((.(.....).)))))).............(((((((..((.((.....)).))(((((((((...................))))))))).........))))))) ( -27.61, z-score = 0.44, R) >droMoj3.scaffold_6496 2664759 78 + 26866924 -----------------------------GGAGGCGUGCCAGAGAG--UGUGUGUGACAAUUUUCUC--AUAUAUCCGAAAUGAAAGAGAAAAUUUAAUUUGCAGGUCACG -----------------------------....(((..(......)--..)))((((((((((((((--.................)))))))))..........))))). ( -14.83, z-score = -0.07, R) >droVir3.scaffold_12875 5855212 87 - 20611582 ---------------------CGGCCGGUCUCUGUG-GGAGGCGUGGCAGUGUGUGACAAUUUUCUC--AUAUAUCCGAAAUGAAAGAGAAAAUUUAAUUUGCAGGUCACG ---------------------..(((((((((....-.))))).)))).....((((((((((((((--.................)))))))))..........))))). ( -24.03, z-score = -1.35, R) >consensus _____________________________GGAGGUGAAAAGCGGGGUGAGAGUGUGACAAUUUUCUC__AUAUAUCCAAAAUGAAAGAGAAAAUUUAAUUUGCAGGUCACG .....................................................((((((((((((((...(((.......)))...)))))))))..........))))). (-10.55 = -10.57 + 0.02)

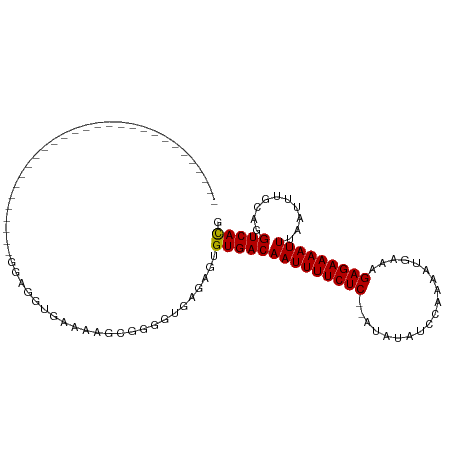
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:10:46 2011