| Sequence ID | dm3.chr2R |
|---|---|
| Location | 5,530,255 – 5,530,347 |
| Length | 92 |
| Max. P | 0.949741 |

| Location | 5,530,255 – 5,530,347 |
|---|---|
| Length | 92 |
| Sequences | 9 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 71.66 |
| Shannon entropy | 0.53685 |
| G+C content | 0.55983 |
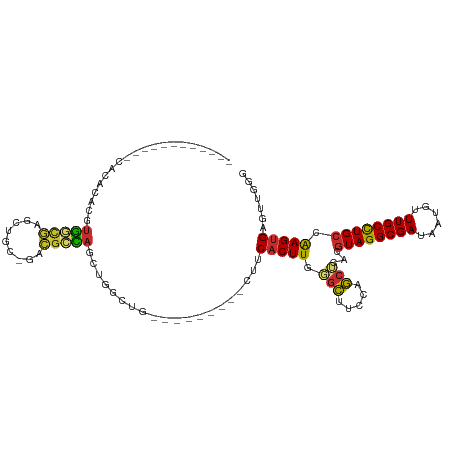
| Mean single sequence MFE | -36.80 |
| Consensus MFE | -18.63 |
| Energy contribution | -18.37 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.949741 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
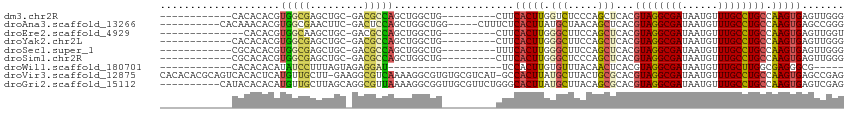
>dm3.chr2R 5530255 92 - 21146708 ------------CACACACGUGGCGAGCUGC-GACGCCAGCUGGCUG---------CUUCACUUGGUCUCCCAGCUCACGUAGGCGAUAAUGUUUGCCUGCCAAGUGAGUUGGG ------------.......(..((.(((((.-.....))))).))..---------)............((((((((((((((((((......))))))))...)))))))))) ( -43.50, z-score = -3.35, R) >droAna3.scaffold_13266 12816304 98 - 19884421 ----------CACAAACACGUGGCGAACUUC-GACUCCAGCUGGCUGG-----CUUUCUCACUUAUGCUAACAGCUCACGUAGGCGAUAAUGUUUGCCUGCCAAGUGAGCCGGG ----------........(.((((..((((.-......(((((..(((-----(............)))).)))))...((((((((......)))))))).))))..)))).) ( -31.90, z-score = -1.47, R) >droEre2.scaffold_4929 17816434 90 + 26641161 --------------CACACGUGGCAAGCUGC-GACGCCAGCUGGCUG---------CUUCACUUGGGCUUCCAGCUCACGUAGGCGAUAAUGUUUGCCUGCCAAGUGAGUUGGU --------------.....(..((.(((((.-.....))))).))..---------).............(((((((((((((((((......))))))))...))))))))). ( -38.70, z-score = -1.94, R) >droYak2.chr2L 18162235 92 - 22324452 ------------CACACACGUGGCGAGCUGC-GACGCCAGCUGGCUG---------CUUCACUUGGGCUUCCAGCUCACGUAGGCGAUAAUGUUUGCCUGCCAAGUGAGUUGGG ------------.......(..((.(((((.-.....))))).))..---------).............(((((((((((((((((......))))))))...))))))))). ( -41.50, z-score = -2.55, R) >droSec1.super_1 3147395 92 - 14215200 ------------CGCACACGUGGCGAGCUGC-GACGCCAGCUGGCUG---------UUUCACUUGGGCUUCCAGCUCACGUAGGCGAUAAUGUUUGCCUGCCAAGUGAGUUGGG ------------.((.((.(((((.(((((.-.....))))).))..---------...))).)).))..(((((((((((((((((......))))))))...))))))))). ( -41.40, z-score = -2.42, R) >droSim1.chr2R 4172057 92 - 19596830 ------------CGCACACGUGGCGAGCUGC-GACGCCAGCUGGCUG---------CUUCACUUGGGCUCCCAGCUCACGUAGGCGAUAAUGUUUGCCUGCCAAGUGAGUUGGG ------------.((....(..((.(((((.-.....))))).))..---------).((....)))).((((((((((((((((((......))))))))...)))))))))) ( -43.70, z-score = -2.60, R) >droWil1.scaffold_180701 3461661 78 - 3904529 ------------CACACACAUAUCCUUUAGUAGAGGAU-------------------UCCACUUGUGUUUACAACUCACGUAGGCGAUAAUGUUUGCUUGGCGAGGGCG----- ------------.(((((...((((((.....))))))-------------------......)))))......(((.(((((((((......)))))).))).)))..----- ( -19.80, z-score = -1.34, R) >droVir3.scaffold_12875 5816932 112 - 20611582 CACACACGCAGUCACACUCAUGUUGCUU-GAAGGCGUCAAAAGGCGUGUGCGUCAU-GCCACUUAUGCUUACUGCGCACGUAGGCGAUAAUGUUUGCCUGCCAAGUGAGCCGAG ..(((.((((((.....(((.......)-))((((((.((..((((((.....)))-)))..))))))))))))))...((((((((......))))))))...)))....... ( -36.80, z-score = -0.10, R) >droGri2.scaffold_15112 746192 104 - 5172618 ----------CAUACACACAUGUUGCUUAGCAGGCGUUAAAAGGCGGUUGCGUUCUGGGCACUUAUGCUUACAGCGCACGUAGGCGAUAAUGUUUGCCUGCCAAGUGAGUCGAG ----------......(((.((.(((...((((.((((....)))).))))(((.((((((....)))))).)))))).((((((((......)))))))))).)))....... ( -33.90, z-score = -0.52, R) >consensus ____________CACACACGUGGCGAGCUGC_GACGCCAGCUGGCUG_________CUUCACUUGGGCUUCCAGCUCACGUAGGCGAUAAUGUUUGCCUGCCAAGUGAGUUGGG ....................(((((.........)))))....................(((((.(((.....)))...((((((((......)))))))).)))))....... (-18.63 = -18.37 + -0.27)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:10:42 2011