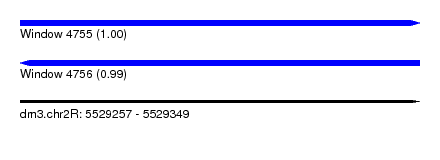
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 5,529,257 – 5,529,349 |
| Length | 92 |
| Max. P | 0.999291 |

| Location | 5,529,257 – 5,529,349 |
|---|---|
| Length | 92 |
| Sequences | 11 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 86.98 |
| Shannon entropy | 0.25485 |
| G+C content | 0.32351 |
| Mean single sequence MFE | -25.53 |
| Consensus MFE | -22.12 |
| Energy contribution | -22.22 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.04 |
| Mean z-score | -4.42 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.77 |
| SVM RNA-class probability | 0.999291 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 5529257 92 + 21146708 -------------------------GGAACCUAGA--CUUUAUUGAUAAAGGCAUAAUUUAUUUGAACAAUCGACAAAACGCUUUUGUCAAUAAAGCCAGAUAAAUAUCUGGCAUUGUA -------------------------..........--.(((((((((((((((.........((((....))))......)))))))))))))))(((((((....)))))))...... ( -26.16, z-score = -5.03, R) >droSim1.chr2R 4171093 92 + 19596830 -------------------------GGAACCUAGA--CUUUAUUGAUAAAGGCAUAAUUUAUUUGAACAAUCGACAAAACGCUUUUGUCAAUAAAGCCAGAUAAAUAUCUGGCAUUGUA -------------------------..........--.(((((((((((((((.........((((....))))......)))))))))))))))(((((((....)))))))...... ( -26.16, z-score = -5.03, R) >droSec1.super_1 3146437 92 + 14215200 -------------------------GGAACCUAGA--CUUUUUUGAUAAAGGCAUAAUUUAUUUGAACAAUCGACAAAACGCUUUUGUCAAUAAAGCCAGAUAAAUAUCUGGCAUUGUA -------------------------..........--.(((.(((((((((((.........((((....))))......))))))))))).)))(((((((....)))))))...... ( -22.26, z-score = -3.48, R) >droYak2.chr2L 18161247 92 + 22324452 -------------------------GGAACCUAGA--CUUUAUUGAUAAAGGCAUAAUUUAUUUGAACAAUCGACAAAACGCUUUUGUCAAUAAAGCCAGAUAAAUAUCUGGCAUUGUA -------------------------..........--.(((((((((((((((.........((((....))))......)))))))))))))))(((((((....)))))))...... ( -26.16, z-score = -5.03, R) >droEre2.scaffold_4929 17815393 92 - 26641161 -------------------------GGAACCUAGA--CUUUAUUGAUAAAGGCAUAAUUUAUUUGAACAAUCGACAAAACGCUUUUGUCAAUAAAGCCAGAUAAAUAUCUGGCAUUGUA -------------------------..........--.(((((((((((((((.........((((....))))......)))))))))))))))(((((((....)))))))...... ( -26.16, z-score = -5.03, R) >droAna3.scaffold_13266 12815360 94 + 19884421 -------------------------CCAACCUAGAGACUUUAUUGAUAAAGGCAUAAUUUAUUUGAACAAUCGACAAAACGCUUUUGUCAAUAAAGCCAGAUAAAUAUCUGGCAUUGUA -------------------------.............(((((((((((((((.........((((....))))......)))))))))))))))(((((((....)))))))...... ( -26.16, z-score = -5.08, R) >dp4.chr3 14158452 91 + 19779522 --------------------------GGAACUAGA--CUUUAUUGAUAAAGGCAUAAUUUAUUUGAACAAUCGACAAAACGCUUUUGUCAAUAAAGCCAGAUAAAUAUCUGGCAUUGUA --------------------------.........--.(((((((((((((((.........((((....))))......)))))))))))))))(((((((....)))))))...... ( -26.16, z-score = -5.30, R) >droPer1.super_4 6002421 91 - 7162766 --------------------------GGAACUAGA--CUUUAUUGAUAAAGGCAUAAUUUAUUUGAACAAUCGACAAAACGCUUUUGUCAAUAAAGCCAGAUAAAUAUCUGGCAUUGUA --------------------------.........--.(((((((((((((((.........((((....))))......)))))))))))))))(((((((....)))))))...... ( -26.16, z-score = -5.30, R) >droWil1.scaffold_180701 3460370 85 + 3904529 ---------------------------------GAA-CUUUAUUGAUAAAGGCAUAAUUUAUUUGAACAAUCGACAAAACGCUAUUGUCAAUAAAGCCAGAUAAAUAUCUGGCAUUGUA ---------------------------------...-.(((((((((((.(((.........((((....))))......))).)))))))))))(((((((....)))))))...... ( -22.96, z-score = -4.59, R) >droVir3.scaffold_12875 5815825 105 + 20611582 ------------GGCGCCACAACACACAGCGCAGAC-UUUUAUUGAUAAGGGCAUAAUUUAUUUGAACAAUCGACAAAACGCUAUUGUCAAUAAAGCCAGAUAAAUAUCUGGCAUUGU- ------------.((((...........))))....-.(((((((((((.(((.........((((....))))......))).)))))))))))(((((((....))))))).....- ( -28.36, z-score = -3.97, R) >droMoj3.scaffold_6496 2623350 118 - 26866924 CUGCUAGACUGGUACAGAAACGCACAGAGCGCAGAC-UUUUAUUGAUAAGGGCAUAAUUUAUUUGAACAAUCGACAAAACGCUAUUGUCAAUAAGGCCAGAUAAAUAUCUUGCAUUGUA .(((.((((((((.......(((.....))).....-.(((((((((((.(((.........((((....))))......))).)))))))))))))))).......))).)))..... ( -24.17, z-score = -0.80, R) >consensus _________________________GGAACCUAGA__CUUUAUUGAUAAAGGCAUAAUUUAUUUGAACAAUCGACAAAACGCUUUUGUCAAUAAAGCCAGAUAAAUAUCUGGCAUUGUA ......................................(((((((((((.(((.........((((....))))......))).)))))))))))(((((((....)))))))...... (-22.12 = -22.22 + 0.10)

| Location | 5,529,257 – 5,529,349 |
|---|---|
| Length | 92 |
| Sequences | 11 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 86.98 |
| Shannon entropy | 0.25485 |
| G+C content | 0.32351 |
| Mean single sequence MFE | -21.94 |
| Consensus MFE | -18.10 |
| Energy contribution | -18.37 |
| Covariance contribution | 0.27 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.65 |
| SVM RNA-class probability | 0.993831 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 5529257 92 - 21146708 UACAAUGCCAGAUAUUUAUCUGGCUUUAUUGACAAAAGCGUUUUGUCGAUUGUUCAAAUAAAUUAUGCCUUUAUCAAUAAAG--UCUAGGUUCC------------------------- ......(((((((....))))(((((((((((.(((.(((((((((.((....))..)))))..)))).))).)))))))))--))..)))...------------------------- ( -21.20, z-score = -3.02, R) >droSim1.chr2R 4171093 92 - 19596830 UACAAUGCCAGAUAUUUAUCUGGCUUUAUUGACAAAAGCGUUUUGUCGAUUGUUCAAAUAAAUUAUGCCUUUAUCAAUAAAG--UCUAGGUUCC------------------------- ......(((((((....))))(((((((((((.(((.(((((((((.((....))..)))))..)))).))).)))))))))--))..)))...------------------------- ( -21.20, z-score = -3.02, R) >droSec1.super_1 3146437 92 - 14215200 UACAAUGCCAGAUAUUUAUCUGGCUUUAUUGACAAAAGCGUUUUGUCGAUUGUUCAAAUAAAUUAUGCCUUUAUCAAAAAAG--UCUAGGUUCC------------------------- .((((((((((((....))))))).....(((((((.....)))))))))))).............((((..((.......)--)..))))...------------------------- ( -19.50, z-score = -2.45, R) >droYak2.chr2L 18161247 92 - 22324452 UACAAUGCCAGAUAUUUAUCUGGCUUUAUUGACAAAAGCGUUUUGUCGAUUGUUCAAAUAAAUUAUGCCUUUAUCAAUAAAG--UCUAGGUUCC------------------------- ......(((((((....))))(((((((((((.(((.(((((((((.((....))..)))))..)))).))).)))))))))--))..)))...------------------------- ( -21.20, z-score = -3.02, R) >droEre2.scaffold_4929 17815393 92 + 26641161 UACAAUGCCAGAUAUUUAUCUGGCUUUAUUGACAAAAGCGUUUUGUCGAUUGUUCAAAUAAAUUAUGCCUUUAUCAAUAAAG--UCUAGGUUCC------------------------- ......(((((((....))))(((((((((((.(((.(((((((((.((....))..)))))..)))).))).)))))))))--))..)))...------------------------- ( -21.20, z-score = -3.02, R) >droAna3.scaffold_13266 12815360 94 - 19884421 UACAAUGCCAGAUAUUUAUCUGGCUUUAUUGACAAAAGCGUUUUGUCGAUUGUUCAAAUAAAUUAUGCCUUUAUCAAUAAAGUCUCUAGGUUGG------------------------- ..(((((((((((....)))))))((((((((.(((.(((((((((.((....))..)))))..)))).))).))))))))........)))).------------------------- ( -23.10, z-score = -3.19, R) >dp4.chr3 14158452 91 - 19779522 UACAAUGCCAGAUAUUUAUCUGGCUUUAUUGACAAAAGCGUUUUGUCGAUUGUUCAAAUAAAUUAUGCCUUUAUCAAUAAAG--UCUAGUUCC-------------------------- ......(((((((....)))))))((((((((.(((.(((((((((.((....))..)))))..)))).))).)))))))).--.........-------------------------- ( -20.90, z-score = -3.44, R) >droPer1.super_4 6002421 91 + 7162766 UACAAUGCCAGAUAUUUAUCUGGCUUUAUUGACAAAAGCGUUUUGUCGAUUGUUCAAAUAAAUUAUGCCUUUAUCAAUAAAG--UCUAGUUCC-------------------------- ......(((((((....)))))))((((((((.(((.(((((((((.((....))..)))))..)))).))).)))))))).--.........-------------------------- ( -20.90, z-score = -3.44, R) >droWil1.scaffold_180701 3460370 85 - 3904529 UACAAUGCCAGAUAUUUAUCUGGCUUUAUUGACAAUAGCGUUUUGUCGAUUGUUCAAAUAAAUUAUGCCUUUAUCAAUAAAG-UUC--------------------------------- ......(((((((....)))))))((((((((.((..(((((((((.((....))..)))))..))))..)).)))))))).-...--------------------------------- ( -20.30, z-score = -3.35, R) >droVir3.scaffold_12875 5815825 105 - 20611582 -ACAAUGCCAGAUAUUUAUCUGGCUUUAUUGACAAUAGCGUUUUGUCGAUUGUUCAAAUAAAUUAUGCCCUUAUCAAUAAAA-GUCUGCGCUGUGUGUUGUGGCGCC------------ -.....(((((((....)))))))((((((((.((..(((((((((.((....))..)))))..))))..)).)))))))).-....((((..(.....)..)))).------------ ( -27.70, z-score = -2.35, R) >droMoj3.scaffold_6496 2623350 118 + 26866924 UACAAUGCAAGAUAUUUAUCUGGCCUUAUUGACAAUAGCGUUUUGUCGAUUGUUCAAAUAAAUUAUGCCCUUAUCAAUAAAA-GUCUGCGCUCUGUGCGUUUCUGUACCAGUCUAGCAG .....(((.((((((((((.((..(..((((((((.......)))))))).)..)).))))))..................(-(...((((.....))))..))......)))).))). ( -24.10, z-score = -1.03, R) >consensus UACAAUGCCAGAUAUUUAUCUGGCUUUAUUGACAAAAGCGUUUUGUCGAUUGUUCAAAUAAAUUAUGCCUUUAUCAAUAAAG__UCUAGGUUCC_________________________ ......(((((((....)))))))((((((((.((..(((((((((.((....))..)))))..))))..)).))))))))...................................... (-18.10 = -18.37 + 0.27)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:10:41 2011