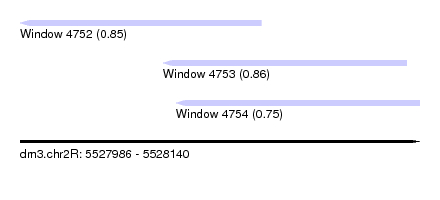
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 5,527,986 – 5,528,140 |
| Length | 154 |
| Max. P | 0.857607 |

| Location | 5,527,986 – 5,528,079 |
|---|---|
| Length | 93 |
| Sequences | 7 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 69.89 |
| Shannon entropy | 0.55150 |
| G+C content | 0.36255 |
| Mean single sequence MFE | -19.27 |
| Consensus MFE | -10.63 |
| Energy contribution | -11.31 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.849737 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
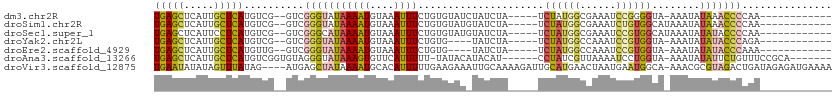
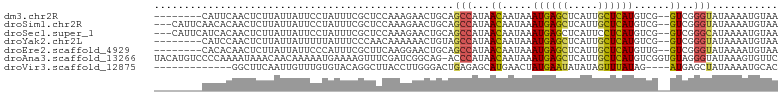
>dm3.chr2R 5527986 93 - 21146708 UGAGCUCAUUGCUCAUGUCG--GUCGGGUAUAAAAUGUAAAUUUCUGUGUAUCUAUCUA-----UCUAUGGCGAAAUCCGGGGUA-AAAUAUAAACCCCAA------------ (((((.....)))))(((((--(..(((((((((((....))))...)))))))..)).-----.....))))......(((((.-........)))))..------------ ( -20.00, z-score = -1.08, R) >droSim1.chr2R 4169823 94 - 19596830 UGAGCUCAUUGCUCAUGUCG--GUCGGGUAUAAAAUGUAAAUUUCUGUGUAUGUAUCUA-----UCUAUGGCGAAAUCUGUGGCAUAAAUAUAAACCCCAA------------ (((((.....))))).....--...((((.....(..((......))..).(((((.((-----(((((((......)))))).))).))))).))))...------------ ( -17.10, z-score = -0.16, R) >droSec1.super_1 3145165 94 - 14215200 UGAGCUCAUUCCUCAUGUCG--GUCGGGCAUAAAAUGUAAAUUUCUGUGUAUGUAUCUA-----UCUAUGGCGAAAUCCGUGGCAUAAAUAUAUACCCCAA------------ ...((((...((.......)--)..))))...................((((((((.((-----(((((((......)))))).))).)))))))).....------------ ( -19.90, z-score = -0.80, R) >droYak2.chr2L 18159998 89 - 22324452 UGAGCUCAUUGCUCAUGUCG--GUCGGGUAUAAAAUGUAAAUUUCUGUG----UAUCUA-----UCUAUGGCCAAAUCCGUGGUA-AAAUAUAUACCCAGA------------ (((((.....))))).....--...(((((((..(..((......))..----).....-----.((((((......))))))..-.....)))))))...------------ ( -20.70, z-score = -1.70, R) >droEre2.scaffold_4929 17814144 89 + 26641161 UGAGCUCAUUGCUCAUGUUG--GUCGGGUAUAAAAUGUAAAUUUCUGUG----UAUCUA-----UCUAUGGCCAAAUCCGUGGUA-AAAUAUAUACCCAAA------------ (((((.....))))).....--...(((((((..(..((......))..----).....-----.((((((......))))))..-.....)))))))...------------ ( -20.70, z-score = -2.19, R) >droAna3.scaffold_13266 12814013 98 - 19884421 UGAGCUCAUUGCUCAUGUCGGUGUAGGGUAUAAAGUGUUCAUUUUU-UAUACAUACAU------CCUAUCGUUAAAAUCCUGGUA-AAAUAUAUUCUGUUUCCGCA------- (((((.....)))))....((((((..((((((((........)))-))))).)))))------)................((..-(((((.....)))))))...------- ( -19.30, z-score = -1.54, R) >droVir3.scaffold_12875 5813979 108 - 20611582 UGAAUAUAUAGUUUAUAG----AUGAGCUAUAAAAUGCACAUUUUUGAAGAAAUUGCAAAAGAUUGCAUGAACUAAUGAAUGGCA-AAACGCGUAGACUGAUAGAGAUGAAAA ......(((((((((...----.))))))))).......((((((((.((....(((((....)))))..............((.-....)).....))..)))))))).... ( -17.20, z-score = -1.21, R) >consensus UGAGCUCAUUGCUCAUGUCG__GUCGGGUAUAAAAUGUAAAUUUCUGUGUAU_UAUCUA_____UCUAUGGCCAAAUCCGUGGUA_AAAUAUAUACCCCAA____________ (((((.....)))))..........(((((((((((....)))).....................((((((......))))))........)))))))............... (-10.63 = -11.31 + 0.68)
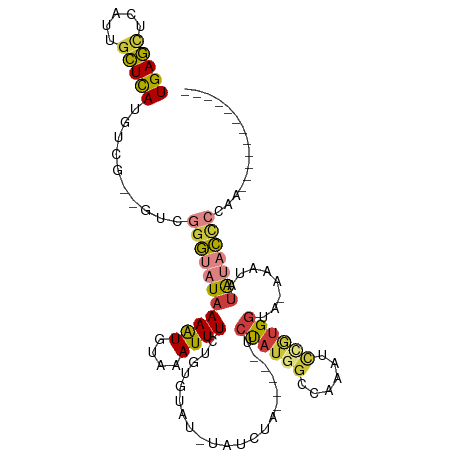
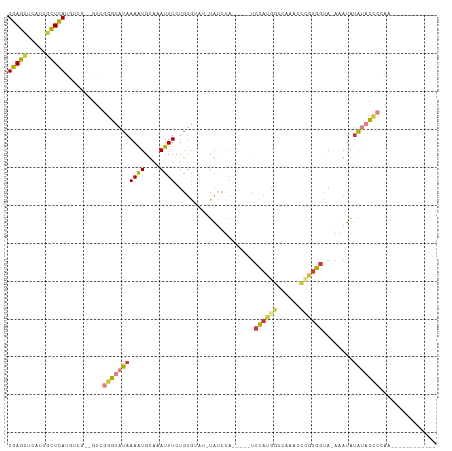
| Location | 5,528,041 – 5,528,135 |
|---|---|
| Length | 94 |
| Sequences | 7 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 67.00 |
| Shannon entropy | 0.65665 |
| G+C content | 0.36988 |
| Mean single sequence MFE | -16.37 |
| Consensus MFE | -8.83 |
| Energy contribution | -8.84 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.44 |
| Mean z-score | -0.81 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.857607 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
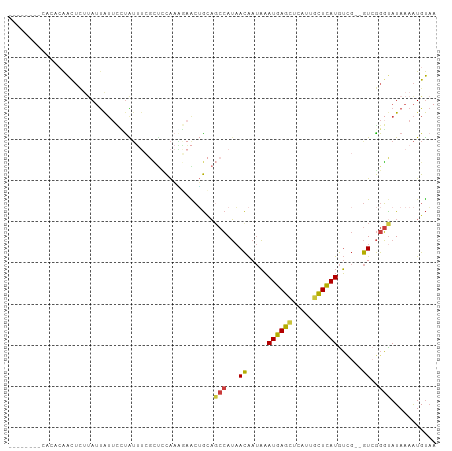
>dm3.chr2R 5528041 94 - 21146708 --------CAUUCAACUCUUAUUAUUCCUAUUUCGCUCCAAAGAACUGCAGCCAUAACAAUAAAUGAGCUCAUUGCUCAUGUCG--GUCGGGUAUAAAAUGUAA --------.............((((.(((.....(((.((......)).)))....((.....((((((.....))))))....--)).))).))))....... ( -15.00, z-score = -0.64, R) >droSim1.chr2R 4169879 99 - 19596830 ---CAUUCAACACAACUCUUAUUAUUCCUAUUUCGCUCCAAAGAACUGCAGCCAUAACAAUAAAUGAGCUCAUUGCUCAUGUCG--GUCGGGUAUAAAAUGUAA ---..................((((.(((.....(((.((......)).)))....((.....((((((.....))))))....--)).))).))))....... ( -15.00, z-score = -0.55, R) >droSec1.super_1 3145221 99 - 14215200 ---CAUUCAUCACAACUCUUAUUAUUCCUAUUUCGCUCCAAAGAACUGCAGCCAUAACAAUAAAUGAGCUCAUUCCUCAUGUCG--GUCGGGCAUAAAAUGUAA ---........(((....................((...........)).(((...((.....(((((.......)))))....--))..)))......))).. ( -11.60, z-score = 0.41, R) >droYak2.chr2L 18160049 94 - 22324452 --------CAUCCAACUCUUAUUAUUUUUAUUUCCCAACAAAAAACUGUAGCCAUAACAAUAAAUGAGCUCAUUGCUCAUGUCG--GUCGGGUAUAAAAUGUAA --------...........(((...((((((..(((.((.......(((.......)))....((((((.....))))))....--)).))).)))))).))). ( -15.70, z-score = -1.69, R) >droEre2.scaffold_4929 17814195 94 + 26641161 --------CACACAACUCUUAUUAUUCCCAUUUCGCUUCAAGGAACUGCAGCCAUAACAAUAAAUGAGCUCAUUGCUCAUGUUG--GUCGGGUAUAAAAUGUAA --------.............((((.(((.....(((.((......)).))).....((((..((((((.....))))))))))--...))).))))....... ( -19.40, z-score = -1.46, R) >droAna3.scaffold_13266 12814071 103 - 19884421 UACAUGUCCCCAAAAUAAACAACAAAAAUGAAAAGUUUCGAUCGGCAG-ACCCAUAACAAUAAAUGAGCUCAUUGCUCAUGUCGGUGUAGGGUAUAAAGUGUUC .((((...(((.................((((....))))((((((.(-........).....((((((.....))))))))))))...)))......)))).. ( -18.30, z-score = -0.55, R) >droVir3.scaffold_12875 5814051 87 - 20611582 -------------GGCUUCAAUUGUUUGUGUACAGGCUUACCUUGGGACUGAGAGCAUGAACUAUGAAUAUAUAGUUUAUAG----AUGAGCUAUAAAAUGCAC -------------.((((((...(((..(((.(((.(((.....))).)))...)))..)))..))))..(((((((((...----.)))))))))....)).. ( -19.60, z-score = -1.18, R) >consensus ________CACACAACUCUUAUUAUUCCUAUUUCGCUCCAAAGAACUGCAGCCAUAACAAUAAAUGAGCUCAUUGCUCAUGUCG__GUCGGGUAUAAAAUGUAA ..................................................(((..........((((((.....))))))..........)))........... ( -8.83 = -8.84 + 0.00)

| Location | 5,528,046 – 5,528,140 |
|---|---|
| Length | 94 |
| Sequences | 7 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 67.47 |
| Shannon entropy | 0.65224 |
| G+C content | 0.36647 |
| Mean single sequence MFE | -15.89 |
| Consensus MFE | -6.87 |
| Energy contribution | -6.84 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.752416 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
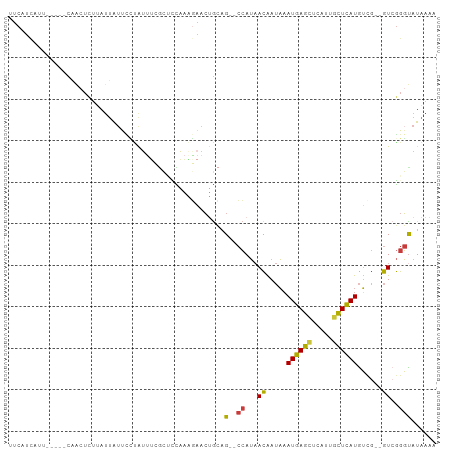
>dm3.chr2R 5528046 94 - 21146708 UUCAUCAUU-----CAACUCUUAUUAUUCCUAUUUCGCUCCAAAGAACUGCAG--CCAUAACAAUAAAUGAGCUCAUUGCUCAUGUCG--GUCGGGUAUAAAA .........-----.........((((.(((.....(((.((......)).))--)....((.....((((((.....))))))....--)).))).)))).. ( -15.00, z-score = -1.10, R) >droSim1.chr2R 4169884 99 - 19596830 AUCAUCAUUCAACACAACUCUUAUUAUUCCUAUUUCGCUCCAAAGAACUGCAG--CCAUAACAAUAAAUGAGCUCAUUGCUCAUGUCG--GUCGGGUAUAAAA .......................((((.(((.....(((.((......)).))--)....((.....((((((.....))))))....--)).))).)))).. ( -15.00, z-score = -1.04, R) >droSec1.super_1 3145226 99 - 14215200 UUCAUCAUUCAUCACAACUCUUAUUAUUCCUAUUUCGCUCCAAAGAACUGCAG--CCAUAACAAUAAAUGAGCUCAUUCCUCAUGUCG--GUCGGGCAUAAAA ....................................((...........)).(--((...((.....(((((.......)))))....--))..)))...... ( -10.80, z-score = 0.17, R) >droYak2.chr2L 18160054 94 - 22324452 UUCAUCAUC-----CAACUCUUAUUAUUUUUAUUUCCCAACAAAAAACUGUAG--CCAUAACAAUAAAUGAGCUCAUUGCUCAUGUCG--GUCGGGUAUAAAA .........-----.............((((((..(((.((.......(((..--.....)))....((((((.....))))))....--)).))).)))))) ( -14.90, z-score = -2.06, R) >droEre2.scaffold_4929 17814200 94 + 26641161 UUCAUCAC-----ACAACUCUUAUUAUUCCCAUUUCGCUUCAAGGAACUGCAG--CCAUAACAAUAAAUGAGCUCAUUGCUCAUGUUG--GUCGGGUAUAAAA ........-----..........((((.(((.....(((.((......)).))--).....((((..((((((.....))))))))))--...))).)))).. ( -19.40, z-score = -2.07, R) >droAna3.scaffold_13266 12814076 103 - 19884421 UUCAUUACAUGUCCCCAAAAUAAACAACAAAAAUGAAAAGUUUCGAUCGGCAGACCCAUAACAAUAAAUGAGCUCAUUGCUCAUGUCGGUGUAGGGUAUAAAG .........(((.(((.................((((....))))((((((.(........).....((((((.....))))))))))))...))).)))... ( -18.20, z-score = -0.95, R) >droVir3.scaffold_12875 5814056 82 - 20611582 ---------------GGCUUCAAUUGUUUGUGUACAGGCUUACCUUGGGACUG--AGAGCAUGAACUAUGAAUAUAUAGUUUAUAG----AUGAGCUAUAAAA ---------------...((((...(((..(((.(((.(((.....))).)))--...)))..)))..))))..(((((((((...----.)))))))))... ( -17.90, z-score = -1.15, R) >consensus UUCAUCAUU_____CAACUCUUAUUAUUCCUAUUUCGCUCCAAAGAACUGCAG__CCAUAACAAUAAAUGAGCUCAUUGCUCAUGUCG__GUCGGGUAUAAAA .......................................................((..........((((((.....))))))..........))....... ( -6.87 = -6.84 + -0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:10:40 2011