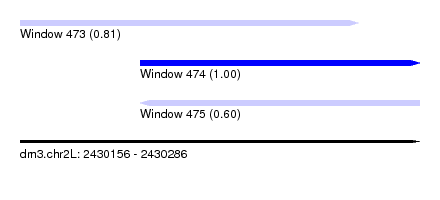
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 2,430,156 – 2,430,286 |
| Length | 130 |
| Max. P | 0.995287 |

| Location | 2,430,156 – 2,430,266 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 91.92 |
| Shannon entropy | 0.11254 |
| G+C content | 0.40909 |
| Mean single sequence MFE | -21.47 |
| Consensus MFE | -17.34 |
| Energy contribution | -17.57 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.812193 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
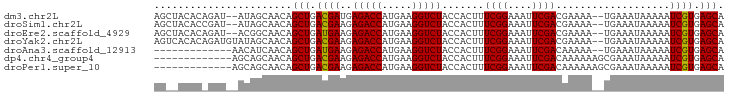
>dm3.chr2L 2430156 110 + 23011544 AACACAUCAGCAAGACAGAGCAGCAAUAUAAUGAGAUAUAGCUACACAGAU--AUAGCAACAGCUGACGAUGAGACCAUGAAGGUCUACCACUUUCGGAAAUUCGACGAAAA ....((((.((........))(((.((((......)))).)))........--.((((....))))..))))(((((.....))))).....(((((.........))))). ( -21.50, z-score = -2.33, R) >droSim1.chr2L 2389342 110 + 22036055 AACACAUCAGCAAGAAAGCGCAGCAAUAUAAUGAGAUAUAGCUACACCGAU--AUAGCAACAGCUGACGAAGAGACCAUGAAGGUCUACCACUUUCGGAAAUUCGACGAAAA .............(((((((((((.((((......)))).((((.......--.))))....)))).))...(((((.....)))))....)))))................ ( -21.60, z-score = -2.16, R) >droYak2.chr2L 2419304 110 + 22324452 AAAACAUCGGCAACACAG--CAGCAAUAUAAUAAGAUAUAGUCACACAGAUGUAUAGCAACAGCUGACGAAGAGACCAUGAAGGUCUACCACUUUCGGAAAUUCGACGAAAA ...((((((....)...(--..((.((((......)))).))..)...))))).((((....)))).((((.(((((.....))))).((......))...))))....... ( -21.30, z-score = -2.11, R) >consensus AACACAUCAGCAAGACAG_GCAGCAAUAUAAUGAGAUAUAGCUACACAGAU__AUAGCAACAGCUGACGAAGAGACCAUGAAGGUCUACCACUUUCGGAAAUUCGACGAAAA ......(((((..............((((......)))).((((..........))))....)))))((((.(((((.....))))).((......))...))))....... (-17.34 = -17.57 + 0.23)


| Location | 2,430,195 – 2,430,286 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 86.15 |
| Shannon entropy | 0.24542 |
| G+C content | 0.41129 |
| Mean single sequence MFE | -21.77 |
| Consensus MFE | -18.16 |
| Energy contribution | -17.96 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.01 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.79 |
| SVM RNA-class probability | 0.995287 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 2430195 91 + 23011544 AGCUACACAGAU--AUAGCAACAGCUGACGAUGAGACCAUGAAGGUCUACCACUUUCGGAAAUUCGACGAAAA--UGAAAUAAAAAUCGUGAGCA .((((.......--.))))....(((.(((((.(((((.....)))))..((.(((((.........))))).--))........))))).))). ( -25.90, z-score = -4.49, R) >droSim1.chr2L 2389381 91 + 22036055 AGCUACACCGAU--AUAGCAACAGCUGACGAAGAGACCAUGAAGGUCUACCACUUUCGGAAAUUCGACGAAAA--UGAAAUAAAAAUCGUGAGCA .((((.......--.))))....(((.((((..(((((.....)))))..((.(((((.........))))).--)).........)))).))). ( -24.90, z-score = -4.19, R) >droEre2.scaffold_4929 2472634 91 + 26641161 AGCUACACAGAU--ACGGCAACAGCUGAUGAAGAGACCAUGAAGGUCUACCACUUUCGGAAAUUCGACGAAAA--UGAAAUAAAAAUCGUGAGCA .(((.(((.(((--.((((....))))......(((((.....)))))..((.(((((.........))))).--))........))))))))). ( -23.20, z-score = -3.19, R) >droYak2.chr2L 2419341 93 + 22324452 AGUCACACAGAUGUAUAGCAACAGCUGACGAAGAGACCAUGAAGGUCUACCACUUUCGGAAAUUCGACGAAAA--UGAAAUAAAAAUCGUGAGCA .(((.....)))...........(((.((((..(((((.....)))))..((.(((((.........))))).--)).........)))).))). ( -21.90, z-score = -2.22, R) >droAna3.scaffold_12913 165235 80 - 441482 -------------AACAUCAACAGCUGAUGAAGAGACCAUGAAGGUCUACCACUUUCGGAAAUUCGACAAAAA--UGAAAUAAAAAUCGUGAGCA -------------..........(((.((((..(((((.....))))).......((((....))))......--...........)))).))). ( -16.70, z-score = -2.11, R) >dp4.chr4_group4 1693276 82 + 6586962 -------------AGCAGCAACAGCUGACGAAGAGACCAUGAAGGUCUACCACUUUCGGAAAUUCGACAAAAAAGCGAAAUAAAAAUCGUGAGCA -------------..........(((.((((..(((((.....))))).((......))...((((.(......))))).......)))).))). ( -19.90, z-score = -2.43, R) >droPer1.super_10 696664 82 + 3432795 -------------AGCAGCAACAGCUGACGAAGAGACCAUGAAGGUCUACCACUUUCGGAAAUUCGACAAAAAAGCGAAAUAAAAAUCGUGAGCA -------------..........(((.((((..(((((.....))))).((......))...((((.(......))))).......)))).))). ( -19.90, z-score = -2.43, R) >consensus AG__ACAC_GAU__ACAGCAACAGCUGACGAAGAGACCAUGAAGGUCUACCACUUUCGGAAAUUCGACGAAAA__UGAAAUAAAAAUCGUGAGCA .......................(((.((((..(((((.....))))).......((((....))))...................)))).))). (-18.16 = -17.96 + -0.20)


| Location | 2,430,195 – 2,430,286 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 86.15 |
| Shannon entropy | 0.24542 |
| G+C content | 0.41129 |
| Mean single sequence MFE | -21.51 |
| Consensus MFE | -13.93 |
| Energy contribution | -14.36 |
| Covariance contribution | 0.43 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.603219 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
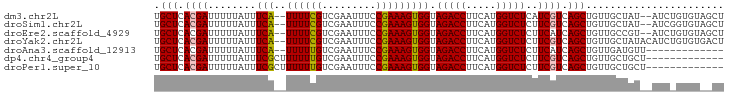
>dm3.chr2L 2430195 91 - 23011544 UGCUCACGAUUUUUAUUUCA--UUUUCGUCGAAUUUCCGAAAGUGGUAGACCUUCAUGGUCUCAUCGUCAGCUGUUGCUAU--AUCUGUGUAGCU .(((.(((((........((--((((((.........))))))))(.(((((.....))))))))))).)))....(((((--(....)))))). ( -27.20, z-score = -4.01, R) >droSim1.chr2L 2389381 91 - 22036055 UGCUCACGAUUUUUAUUUCA--UUUUCGUCGAAUUUCCGAAAGUGGUAGACCUUCAUGGUCUCUUCGUCAGCUGUUGCUAU--AUCGGUGUAGCU .(((.((((........(((--((((((.........))))))))).(((((.....)))))..)))).)))....(((((--(....)))))). ( -26.40, z-score = -3.41, R) >droEre2.scaffold_4929 2472634 91 - 26641161 UGCUCACGAUUUUUAUUUCA--UUUUCGUCGAAUUUCCGAAAGUGGUAGACCUUCAUGGUCUCUUCAUCAGCUGUUGCCGU--AUCUGUGUAGCU .(((((((((.......(((--((((((.........))))))))).(((((.....)))))....))).((....))...--....))).))). ( -21.60, z-score = -1.99, R) >droYak2.chr2L 2419341 93 - 22324452 UGCUCACGAUUUUUAUUUCA--UUUUCGUCGAAUUUCCGAAAGUGGUAGACCUUCAUGGUCUCUUCGUCAGCUGUUGCUAUACAUCUGUGUGACU .(((.((((........(((--((((((.........))))))))).(((((.....)))))..)))).))).((..(...........)..)). ( -23.70, z-score = -2.32, R) >droAna3.scaffold_12913 165235 80 + 441482 UGCUCACGAUUUUUAUUUCA--UUUUUGUCGAAUUUCCGAAAGUGGUAGACCUUCAUGGUCUCUUCAUCAGCUGUUGAUGUU------------- (((.(((((........)).--......(((......)))..))))))((((.....))))....((((((...))))))..------------- ( -14.70, z-score = -0.38, R) >dp4.chr4_group4 1693276 82 - 6586962 UGCUCACGAUUUUUAUUUCGCUUUUUUGUCGAAUUUCCGAAAGUGGUAGACCUUCAUGGUCUCUUCGUCAGCUGUUGCUGCU------------- .(((.((((........((((((.....(((......))))))))).(((((.....)))))..)))).)))..........------------- ( -18.50, z-score = -1.13, R) >droPer1.super_10 696664 82 - 3432795 UGCUCACGAUUUUUAUUUCGCUUUUUUGUCGAAUUUCCGAAAGUGGUAGACCUUCAUGGUCUCUUCGUCAGCUGUUGCUGCU------------- .(((.((((........((((((.....(((......))))))))).(((((.....)))))..)))).)))..........------------- ( -18.50, z-score = -1.13, R) >consensus UGCUCACGAUUUUUAUUUCA__UUUUCGUCGAAUUUCCGAAAGUGGUAGACCUUCAUGGUCUCUUCGUCAGCUGUUGCUGU__AUC_GUGU__CU .(((.((((...................(((......)))....((.(((((.....))))))))))).)))....................... (-13.93 = -14.36 + 0.43)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:11:14 2011