| Sequence ID | dm3.chr2R |
|---|---|
| Location | 5,450,070 – 5,450,186 |
| Length | 116 |
| Max. P | 0.699077 |

| Location | 5,450,070 – 5,450,186 |
|---|---|
| Length | 116 |
| Sequences | 15 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 68.95 |
| Shannon entropy | 0.72261 |
| G+C content | 0.55230 |
| Mean single sequence MFE | -41.43 |
| Consensus MFE | -14.81 |
| Energy contribution | -14.05 |
| Covariance contribution | -0.76 |
| Combinations/Pair | 2.18 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.699077 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
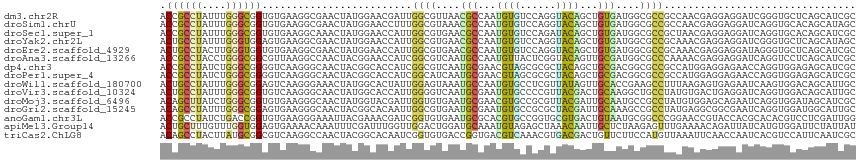
>dm3.chr2R 5450070 116 + 21146708 ACCGCCUAUUUGGGCGGUGUGAAGGCGAACUAUGGAACGAUUGGCGUUAACGCCAAUGUGUCCAGGUACAGCUGUGAUGGCGCCGCCAACGAGGAGGAUCGGGUGCUCAGCAUCGC .((.(((.....((((((((.(.(((..(((.((((...(((((((....)))))))...)))))))...)))....).))))))))....))).))....(((((...))))).. ( -48.70, z-score = -2.20, R) >droSim1.chrU 15111180 116 - 15797150 ACCGCCUAUUUGGGCGGUGUGAAGGCGAACUAUGGAACCUUUGGCGUAAACGCCAAUGUGUCCAGGUACAGCUGUGAUGGCGCCGCCAACGAGGAGGAUCAGGUGCACAGCAUAGC (((((((....)))))))........(.(((.((((..(.((((((....)))))).)..))))))).).((((((.(((((((.((........))....))))).)).)))))) ( -47.60, z-score = -2.39, R) >droSec1.super_1 3064808 116 + 14215200 ACCGCCUAUUUGGGCGGUGUGAAGGCAAACUAUGGAACCAUUGGCGUGAACGCCAAUGUGUCCAGAUACAGCUGUGAUGGCGCCGCUAACGAGGAGGAUCAGGUGCACAGCAUCGC .((.(((.....((((((((.(.(((......((((..((((((((....))))))))..))))......)))....).))))))))....))).))....(((((...))))).. ( -50.70, z-score = -3.62, R) >droYak2.chr2L 18081577 116 + 22324452 ACUGCCUAUUUGGGUGGAGUGAAGGCGAACUAUGGAACCAUUGGCGUGAACGCCAAUGUGUCCAGGUACAGCUGUGAUGGCGCCGCAAACGAGGAGGAUCGGGUGCUCAGCAUAGC .(..(((....)))..)..(((.((((.(((.((((..((((((((....))))))))..))))))).).(((.....))))))(((..(((......)))..))))))....... ( -46.40, z-score = -2.63, R) >droEre2.scaffold_4929 17734786 116 - 26641161 ACUGCCUACUUGGGUGGUGUGAAGGCGAACUAUGGAACCAUUGGCGUGAACGCCAAUGUGUCCAGGUACAGCUGUGAUGGCGCCGCAAACGAGGAGGAUAGGGUGCUCAGCAUCGC .((((((.((((.(((((((.(.(((..(((.((((..((((((((....))))))))..)))))))...)))....).)))))))...)))).))).)))(((((...))))).. ( -51.90, z-score = -3.75, R) >droAna3.scaffold_13266 19710617 116 - 19884421 ACCGCCUACCUGGGCGGCGUUAAGGCCAACUACGGAACCAUCGGCGUCAAUGCCAAUGUUACUCGGUACAGUUGCGAUGGCGCCCAAAACGAGGAGGAUCAGGUCCUCAGCAUCGC .((((((....))))(((......)))......)).......(((((((.(((...(((........)))...))).))))))).....(((.((((((...))))))....))). ( -39.50, z-score = -0.38, R) >dp4.chr3 15891036 116 + 19779522 ACCGCCUAUCUGGGCGGGGUCAAGGGCAACUACGGCACCAUCGGCGUCAAUGCGAACGUAGCGCGCUACAGCUGCGACGGCGCCGCCAUGGAGGAGAACCAGGUGGAGAGCAUCGC .((((((.(((((((((.(((....(((...(((.(......).)))...)))...((((((........))))))..))).))))).))))((....)))))))).......... ( -45.60, z-score = -0.13, R) >droPer1.super_4 6291790 116 - 7162766 ACCGCCUAUCUGGGCGGGGUCAAGGGCAACUACGGCACCAUCGGCAUCAAUGCGAACGUAGCGCGCUACAGCUGCGACGGCGCCGCCAUGGAGGAGAACCAGGUGGAGAGCAUCGC .((((((....))))))((((..((....))...).)))............((((.((((((........))))))...((.(((((.(((.......))))))))...)).)))) ( -44.30, z-score = -0.23, R) >droWil1.scaffold_180700 3314310 116 + 6630534 ACUGCCUAUUUGGGCGGAGUCAAGGGAAACUAUGGCACUAUUGGAGUAAAUGCCAAUGUGCCUCGUUAUAGUUGCACCGAAGCCUUUAAGAGUGAGAAUCAAGUGGACAGCAUUGC .((((((....)))))).(((..((..(((((((((((.(((((........)))))))))).....))))))...)).....(((.......))).........)))........ ( -32.00, z-score = -0.47, R) >droVir3.scaffold_10324 243322 116 + 1288806 ACUGCCUAUUUGGGCGGUGUCAAGGGCAACUAUGGCACCAUUGGGGUCAAUGCGAAUGUGCCCCGUUACGACUGCAAGGCUGCCUAUGUGACUGAGGAUCAGGUGGACAGCAUUGC ...((((....))))(((((((.((....)).)))))))...(((((..((....))..))))).........((((.((((.((((.((((....).))).)))).)))).)))) ( -45.90, z-score = -1.70, R) >droMoj3.scaffold_6496 2061512 116 + 26866924 ACAGCUUAUCUGGGCGGUGUGAAGGGCAACUAUGGUACGAUUGGUGUGAAUGCGAACGUGCCGCGUUACGAUUGCAAUGCCGCCUAUGUGGAGCAGAAUCAGGUGGAUAGCAUCGC ...((((...((((((((((...((....))..((((((.((.((......)).))))))))(((.......))).))))))))))....)))).......((((.....)))).. ( -38.10, z-score = -0.25, R) >droGri2.scaffold_15245 2948443 116 - 18325388 ACAGCCUAUUUGGGCGGAGUGAAGGGCAACUACGGCACAAUUGGCGUGAAUGCGAAUGUGCCGCGCUACGAUUGCAAAGCCGCCUAUGAGGCGGCGAAUCAGGUGGAUGGCAUUGC ...((((((((((.((.((((..((....)).(((((((.((.(((....))).))))))))))))).))........((((((.....))))))...)))))))...)))..... ( -44.10, z-score = -1.43, R) >anoGam1.chr3L 19901483 116 + 41284009 ACCGCCUAUCUGACCGGUGUGAAGGGAAAUUACGAAACGAUCGGUGUGAAUGCGCACGUGCCGGUGCGUGACUGUAAUGCGGCCCGGAACCGUACCACGCACACGUCCUCGAUUGG ....((.(((.(((..(((((..(((..((((((..((((((((..((........))..))))).)))...))))))....)))((.......)).)))))..)))...))).)) ( -35.50, z-score = 1.10, R) >apiMel3.Group14 2261431 116 + 8318479 ACUGCUUUGUUUGGUGGAGUGAAAACAAAUUUCGAUUUGGUUGGACUGGAUGCAAAUGUAGAGCUAAACAAUUGCUCUAAGAGUUUGAAAACAGAUUAUCAUGUGGAUUCUAUUAU ...........(((((((((....)).....((.((.((((....(((....(((((.((((((.........))))))...)))))....)))...)))).)).)).))))))). ( -26.10, z-score = -1.22, R) >triCas2.ChLG8 13534325 116 + 15773733 ACAGCCUACUUAUGCGGCGUCAAGGCCAACUACGGCACAAUCGGUGUGACCGGUGACGUCAAACGUGACGACUGUUCUUCCAUGUUAAAUUCAACCAAUCACGUCCAUUCAAUCGC .............((((((((...(((......)))...(((((.....)))))))))))..((((((.((....))......(((......)))...))))))..........)) ( -25.00, z-score = 0.87, R) >consensus ACCGCCUAUUUGGGCGGUGUGAAGGCCAACUAUGGAACCAUUGGCGUGAAUGCCAAUGUGCCCCGGUACAGCUGCGAUGGCGCCGCUAACGAGGAGAAUCAGGUGCACAGCAUCGC .((((((....)))))).........................((((....(((...((((......))))...)))....))))................................ (-14.81 = -14.05 + -0.76)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:10:29 2011