| Sequence ID | dm3.chr2R |
|---|---|
| Location | 5,441,245 – 5,441,352 |
| Length | 107 |
| Max. P | 0.999049 |

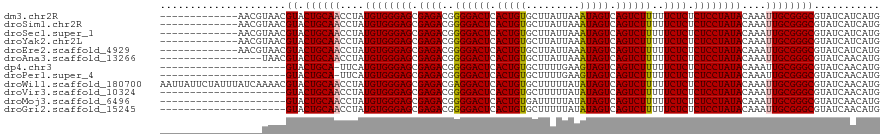
| Location | 5,441,245 – 5,441,352 |
|---|---|
| Length | 107 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.32 |
| Shannon entropy | 0.18267 |
| G+C content | 0.44780 |
| Mean single sequence MFE | -38.51 |
| Consensus MFE | -36.00 |
| Energy contribution | -35.95 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.06 |
| Mean z-score | -5.13 |
| Structure conservation index | 0.93 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.62 |
| SVM RNA-class probability | 0.999049 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
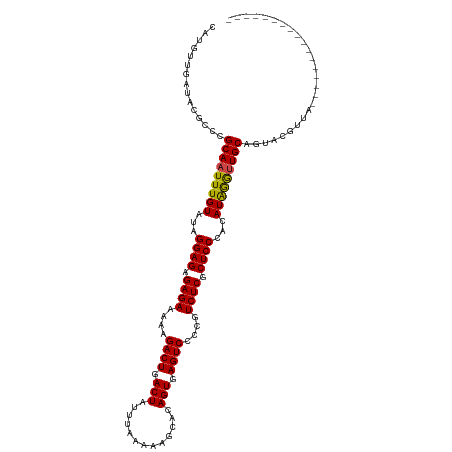
>dm3.chr2R 5441245 107 + 21146708 -------------AACGUAACGUACUGCAACCUAUGUGGGAGCGAGACGGGGACUCACUGUGCUUAUUAAAUAGUCAGUCUUUUUCUCUCUCCUAUACAAAUUGCGGGCGUAUCAUCAUG -------------......((((.((((((....((((((((.((((..((((((.(((((.........))))).))))))..)))).))))))))....))))))))))......... ( -40.30, z-score = -5.46, R) >droSim1.chr2R 4072815 107 + 19596830 -------------AACGUAACGUACUGCAACCUAUGUGGGAGCGAGACGGGGACUCACUGUGCUUAUUAAAUAGUCAGUCUUUUUCUCUCUCCUAUACAAAUUGCGGGCGUAUCAUCAUG -------------......((((.((((((....((((((((.((((..((((((.(((((.........))))).))))))..)))).))))))))....))))))))))......... ( -40.30, z-score = -5.46, R) >droSec1.super_1 3055909 107 + 14215200 -------------AACGUAACGUACUGCAACCUAUGUGGGAGCGAGACGGGGACUCACUGUGCUUAUUAAAUAGUCAGUCUUUUUCUCUCUCCUAUACAAAUUGCGGGCGUAUCAUCAUG -------------......((((.((((((....((((((((.((((..((((((.(((((.........))))).))))))..)))).))))))))....))))))))))......... ( -40.30, z-score = -5.46, R) >droYak2.chr2L 18072260 107 + 22324452 -------------AACGUAACGUACUGCAACCUAUGUGGGAGCGAGACGGGGACUCACUGUGCUUAUUAAAUAGUCAGUCUUUUUCUCUCUCCUAUACAAAUUGCGGGCGUAUCAUCAUG -------------......((((.((((((....((((((((.((((..((((((.(((((.........))))).))))))..)))).))))))))....))))))))))......... ( -40.30, z-score = -5.46, R) >droEre2.scaffold_4929 17725161 107 - 26641161 -------------AACGUAACGUACUGCAACCUAUGUGGGAGCGAGACGGGGACUCACUGUGCUUAUUAAAUAGUCAGUCUUUUUCUCUCUCCUAUACAAAUUGCGGGCGUAUCAUCAUG -------------......((((.((((((....((((((((.((((..((((((.(((((.........))))).))))))..)))).))))))))....))))))))))......... ( -40.30, z-score = -5.46, R) >droAna3.scaffold_13266 19700372 103 - 19884421 -----------------UAACGUACUGCAACCUAUGUGGGAGCGAGACGGGGACUCACUGUGCUUAUUAAAUAGUCAGUCUUUUUCUCUCUCCUAUACAAAUUGCGGGCGUAUCAACAUG -----------------..((((.((((((....((((((((.((((..((((((.(((((.........))))).))))))..)))).))))))))....))))))))))......... ( -40.30, z-score = -5.98, R) >dp4.chr3 15883113 98 + 19779522 ---------------------GUACUGCA-UUCAUGUGGGAGCGAGACGGGGACUCACUGUGCUUUUGAAGUAGUCAGUCUUUUUCUCUCUCCUAUACAAAUUGCGGGCGUAUCAACAUG ---------------------((.(((((-....((((((((.((((..((((((.((((..(....)...)))).))))))..)))).)))))))).....)))))))........... ( -34.20, z-score = -3.43, R) >droPer1.super_4 6283929 98 - 7162766 ---------------------GUACUGCA-UUCAUGUGGGAGCGAGACGGGGACUCACUGUGCUUUUGAAGUAGUCAGUCUUUUUCUCUCUCCUAUACAAAUUGCGGGCGUAUCAACAUG ---------------------((.(((((-....((((((((.((((..((((((.((((..(....)...)))).))))))..)))).)))))))).....)))))))........... ( -34.20, z-score = -3.43, R) >droWil1.scaffold_180700 3302329 120 + 6630534 AAUUAUUCUAUUUAUCAAAACGUACUGCAACCUAUGUGGGAGCGAGACGAGGACUCACUGUGCUUUUUAUAUAGUCAGUCUUUUUCUCUCUCCUAUACAAAUUGCGGGCGUAUCAACAUG ...................((((.((((((....((((((((.((((.(((((((.((((((.......)))))).))))))).)))).))))))))....))))))))))......... ( -40.90, z-score = -6.17, R) >droVir3.scaffold_10324 225314 99 + 1288806 ---------------------GUACUGCAACCUAUGUGGGAGCGAGACGGGGACUCACUGUGCUUUUUAUAUAGUCAGUCUUUUUCUCUCUCCUAUACAAAUUGCGGGCGUAUCAACAUG ---------------------((.((((((....((((((((.((((..((((((.((((((.......)))))).))))))..)))).))))))))....))))))))........... ( -37.10, z-score = -5.13, R) >droMoj3.scaffold_6496 2042843 99 + 26866924 ---------------------GUACUGCAACCUAUGUGGGAGCGAGACGGGGACUCACUGUGAUUUUUAUAUAGUCAGUCUUUUUCUCUCUCCUAUACAAAUUGCGGGCGUAUCAACAUG ---------------------((.((((((....((((((((.((((..((((((.((((((.......)))))).))))))..)))).))))))))....))))))))........... ( -36.80, z-score = -5.06, R) >droGri2.scaffold_15245 2923632 99 - 18325388 ---------------------GUACUGCAACCUAUGUGGGAGCGAGACGGGGACUCACUGUGCUUUUUAUAUAGUCAGUCUUUUUCUCUCUCCUAUACAAAUUGCGGGCGUAUCAACAUG ---------------------((.((((((....((((((((.((((..((((((.((((((.......)))))).))))))..)))).))))))))....))))))))........... ( -37.10, z-score = -5.13, R) >consensus _________________UAACGUACUGCAACCUAUGUGGGAGCGAGACGGGGACUCACUGUGCUUAUUAAAUAGUCAGUCUUUUUCUCUCUCCUAUACAAAUUGCGGGCGUAUCAACAUG .....................((.((((((....((((((((.((((..((((((.(((((.........))))).))))))..)))).))))))))....))))))))........... (-36.00 = -35.95 + -0.05)

| Location | 5,441,245 – 5,441,352 |
|---|---|
| Length | 107 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.32 |
| Shannon entropy | 0.18267 |
| G+C content | 0.44780 |
| Mean single sequence MFE | -29.41 |
| Consensus MFE | -28.25 |
| Energy contribution | -28.00 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.20 |
| Structure conservation index | 0.96 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.22 |
| SVM RNA-class probability | 0.997958 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 5441245 107 - 21146708 CAUGAUGAUACGCCCGCAAUUUGUAUAGGAGAGAGAAAAAGACUGACUAUUUAAUAAGCACAGUGAGUCCCCGUCUCGCUCCCACAUAGGUUGCAGUACGUUACGUU------------- ..(((((.(((....(((((((((...((((.((((....((((.(((.............))).))))....)))).))))...))))))))).))))))))....------------- ( -31.52, z-score = -3.47, R) >droSim1.chr2R 4072815 107 - 19596830 CAUGAUGAUACGCCCGCAAUUUGUAUAGGAGAGAGAAAAAGACUGACUAUUUAAUAAGCACAGUGAGUCCCCGUCUCGCUCCCACAUAGGUUGCAGUACGUUACGUU------------- ..(((((.(((....(((((((((...((((.((((....((((.(((.............))).))))....)))).))))...))))))))).))))))))....------------- ( -31.52, z-score = -3.47, R) >droSec1.super_1 3055909 107 - 14215200 CAUGAUGAUACGCCCGCAAUUUGUAUAGGAGAGAGAAAAAGACUGACUAUUUAAUAAGCACAGUGAGUCCCCGUCUCGCUCCCACAUAGGUUGCAGUACGUUACGUU------------- ..(((((.(((....(((((((((...((((.((((....((((.(((.............))).))))....)))).))))...))))))))).))))))))....------------- ( -31.52, z-score = -3.47, R) >droYak2.chr2L 18072260 107 - 22324452 CAUGAUGAUACGCCCGCAAUUUGUAUAGGAGAGAGAAAAAGACUGACUAUUUAAUAAGCACAGUGAGUCCCCGUCUCGCUCCCACAUAGGUUGCAGUACGUUACGUU------------- ..(((((.(((....(((((((((...((((.((((....((((.(((.............))).))))....)))).))))...))))))))).))))))))....------------- ( -31.52, z-score = -3.47, R) >droEre2.scaffold_4929 17725161 107 + 26641161 CAUGAUGAUACGCCCGCAAUUUGUAUAGGAGAGAGAAAAAGACUGACUAUUUAAUAAGCACAGUGAGUCCCCGUCUCGCUCCCACAUAGGUUGCAGUACGUUACGUU------------- ..(((((.(((....(((((((((...((((.((((....((((.(((.............))).))))....)))).))))...))))))))).))))))))....------------- ( -31.52, z-score = -3.47, R) >droAna3.scaffold_13266 19700372 103 + 19884421 CAUGUUGAUACGCCCGCAAUUUGUAUAGGAGAGAGAAAAAGACUGACUAUUUAAUAAGCACAGUGAGUCCCCGUCUCGCUCCCACAUAGGUUGCAGUACGUUA----------------- .........(((..((((((((((...((((.((((....((((.(((.............))).))))....)))).))))...))))))))).)..)))..----------------- ( -29.42, z-score = -3.10, R) >dp4.chr3 15883113 98 - 19779522 CAUGUUGAUACGCCCGCAAUUUGUAUAGGAGAGAGAAAAAGACUGACUACUUCAAAAGCACAGUGAGUCCCCGUCUCGCUCCCACAUGAA-UGCAGUAC--------------------- ...............(((.((..(...((((.((((....((((.(((.............))).))))....)))).))))...)..))-))).....--------------------- ( -23.32, z-score = -1.74, R) >droPer1.super_4 6283929 98 + 7162766 CAUGUUGAUACGCCCGCAAUUUGUAUAGGAGAGAGAAAAAGACUGACUACUUCAAAAGCACAGUGAGUCCCCGUCUCGCUCCCACAUGAA-UGCAGUAC--------------------- ...............(((.((..(...((((.((((....((((.(((.............))).))))....)))).))))...)..))-))).....--------------------- ( -23.32, z-score = -1.74, R) >droWil1.scaffold_180700 3302329 120 - 6630534 CAUGUUGAUACGCCCGCAAUUUGUAUAGGAGAGAGAAAAAGACUGACUAUAUAAAAAGCACAGUGAGUCCUCGUCUCGCUCCCACAUAGGUUGCAGUACGUUUUGAUAAAUAGAAUAAUU ..(((..(.(((..((((((((((...((((.((((....((((.(((.............))).))))....)))).))))...))))))))).)..))).)..)))............ ( -34.02, z-score = -3.84, R) >droVir3.scaffold_10324 225314 99 - 1288806 CAUGUUGAUACGCCCGCAAUUUGUAUAGGAGAGAGAAAAAGACUGACUAUAUAAAAAGCACAGUGAGUCCCCGUCUCGCUCCCACAUAGGUUGCAGUAC--------------------- ...............(((((((((...((((.((((....((((.(((.............))).))))....)))).))))...))))))))).....--------------------- ( -28.42, z-score = -3.49, R) >droMoj3.scaffold_6496 2042843 99 - 26866924 CAUGUUGAUACGCCCGCAAUUUGUAUAGGAGAGAGAAAAAGACUGACUAUAUAAAAAUCACAGUGAGUCCCCGUCUCGCUCCCACAUAGGUUGCAGUAC--------------------- ...............(((((((((...((((.((((....((((.(((.............))).))))....)))).))))...))))))))).....--------------------- ( -28.42, z-score = -3.64, R) >droGri2.scaffold_15245 2923632 99 + 18325388 CAUGUUGAUACGCCCGCAAUUUGUAUAGGAGAGAGAAAAAGACUGACUAUAUAAAAAGCACAGUGAGUCCCCGUCUCGCUCCCACAUAGGUUGCAGUAC--------------------- ...............(((((((((...((((.((((....((((.(((.............))).))))....)))).))))...))))))))).....--------------------- ( -28.42, z-score = -3.49, R) >consensus CAUGUUGAUACGCCCGCAAUUUGUAUAGGAGAGAGAAAAAGACUGACUAUUUAAAAAGCACAGUGAGUCCCCGUCUCGCUCCCACAUAGGUUGCAGUACGUUA_________________ ...............(((((((((...((((.((((....((((.(((.............))).))))....)))).))))...))))))))).......................... (-28.25 = -28.00 + -0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:10:28 2011