| Sequence ID | dm3.chr2R |
|---|---|
| Location | 5,434,521 – 5,434,633 |
| Length | 112 |
| Max. P | 0.661316 |

| Location | 5,434,521 – 5,434,633 |
|---|---|
| Length | 112 |
| Sequences | 7 |
| Columns | 124 |
| Reading direction | reverse |
| Mean pairwise identity | 67.08 |
| Shannon entropy | 0.62169 |
| G+C content | 0.46421 |
| Mean single sequence MFE | -23.88 |
| Consensus MFE | -10.98 |
| Energy contribution | -13.14 |
| Covariance contribution | 2.17 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.04 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.661316 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
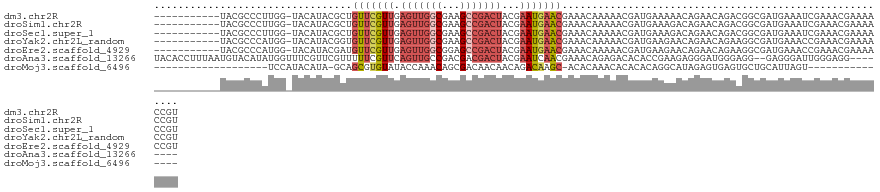
>dm3.chr2R 5434521 112 - 21146708 -----------UACGCCCUUGG-UACAUACGCUGUUCGUUGAGUUGGCGAAGCCGACUACGAAUGAACGAAACAAAAACGAUGAAAAACAGAACAGACGGCGAUGAAAUCGAAACGAAAACCGU -----------.........((-(.(((.((((((..(((.(((((((...))))))).((......))......................)))..)))))))))...(((...)))..))).. ( -25.50, z-score = -1.55, R) >droSim1.chr2R 4065986 112 - 19596830 -----------UACGCCCUUGG-UACAUACGCUGUUCGUUGAGUUGGCGAAGCCGACUACGAAUGAACGAAACAAAAACGAUGAAAGACAGAACAGACGGCGAUGAAAUCGAAACGAAAACCGU -----------.........((-(.(((.((((((..(((.(((((((...))))))).((......))......................)))..)))))))))...(((...)))..))).. ( -25.50, z-score = -1.39, R) >droSec1.super_1 3049210 112 - 14215200 -----------UACGCCCUUGG-UACAUACGCUGUUCGUUGAGUUGGCGAAGCCGACUACGAAUGAACGAAACAAAAACGAUGAAAGACAGAACAGACGGCGAUGAAAUCGAAACGAAAACCGU -----------.........((-(.(((.((((((..(((.(((((((...))))))).((......))......................)))..)))))))))...(((...)))..))).. ( -25.50, z-score = -1.39, R) >droYak2.chr2L_random 4057927 112 - 4064425 -----------UACGCCCAUGG-UACAUACGGUGUUCGUUGAGUUGGCGAAGCCGACUACGAAUGAACGAAACAAAAACGAUGAAGAACAGAACAGAAGGCGAUGAAACCGAAACGAAAACCGU -----------..(((((((.(-(........((((((((.(((((((...)))))))...))))))))........)).))).....(......)..))))...........(((.....))) ( -25.19, z-score = -1.60, R) >droEre2.scaffold_4929 17718266 112 + 26641161 -----------UACGCCCAUGG-UACAUACGAUGUUCGUUGAGUUGGCGGAGCCGACUACGAAUGAACGAAACAAAAACGAUGAAGAACAGAACAGAAGGCGAUGAAACCGAAACGAAAACCGU -----------..(((((((.(-(....)).)))((((((((((((((...))))))).((......)).........))))))).............))))...........(((.....))) ( -24.20, z-score = -1.65, R) >droAna3.scaffold_13266 19694529 114 + 19884421 UACACCUUUAAUGUACAUAUGGUUUCGUUCGUUUUUCGUUCAGUUGCCGACGACGACUACGAAUCAACGAAACAGAGACACACCGAAGAGGGAUGGGAGG--GAGGGAUUGGGAGG-------- ....(((((......(((.(((((((.((((((.(((((...((((....))))....)))))..))))))...))))).))((.....)).))).....--))))).........-------- ( -27.70, z-score = -0.05, R) >droMoj3.scaffold_6496 2033173 88 - 26866924 -------------------UCCAUACAUA-GCAGCGUGUAUACCAAACAGCGACAACAACAGACAAGC-ACACAAACACACACAGGCAUAGAGUGAGUGCUGCAUUAGU--------------- -------------------..........-(((((((((..........((...............))-))))..((.(((.(.......).))).)))))))......--------------- ( -13.56, z-score = 0.34, R) >consensus ___________UACGCCCAUGG_UACAUACGCUGUUCGUUGAGUUGGCGAAGCCGACUACGAAUGAACGAAACAAAAACGAUGAAAAACAGAACAGAAGGCGAUGAAAUCGAAACGAAAACCGU .............((((..((....)).....((((((((.(((((((...)))))))...)))))))).............................))))...................... (-10.98 = -13.14 + 2.17)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:10:26 2011