
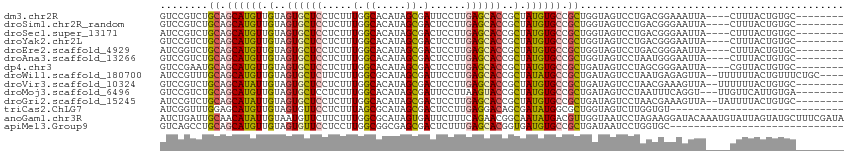
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 5,414,917 – 5,415,019 |
| Length | 102 |
| Max. P | 0.881780 |

| Location | 5,414,917 – 5,415,019 |
|---|---|
| Length | 102 |
| Sequences | 14 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 79.70 |
| Shannon entropy | 0.44150 |
| G+C content | 0.50011 |
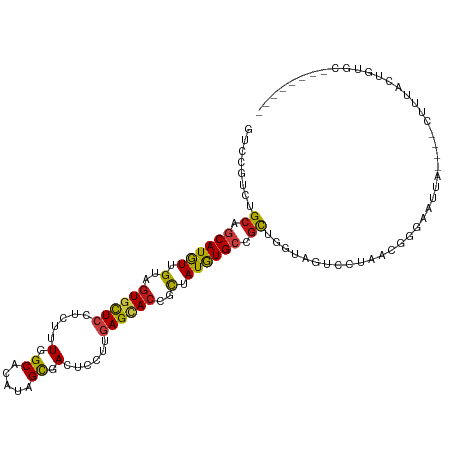
| Mean single sequence MFE | -32.32 |
| Consensus MFE | -18.78 |
| Energy contribution | -18.59 |
| Covariance contribution | -0.18 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.881780 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 5414917 102 - 21146708 GUCCGUCUGCAGCAUGUUGUAGUGCUCCUCUUUGGCACAUAGCGAUUCCUUGAGCACCGCUAUGUGCCGCUGGUAGUCCUGACGGAAAUUA----CUUUACUGUGC-------- .((((((.(((((((......))))).......(((((((((((.............))))))))))))).((....)).)))))).....----...........-------- ( -36.32, z-score = -2.71, R) >droSim1.chr2R_random 1718595 102 - 2996586 GUCCGUCUGCAGCAUGUUGUAGUGCUCCUCUUUGGCACAUAGCGACUCCUUGAGCACCGCUAUGUGCCGCUGGUAGUCCUGACGGGAAUUA----CUUUACUGUGC-------- .((((((.(((((((......))))).......(((((((((((.(((...)))...))))))))))))).((....)).)))))).....----...........-------- ( -36.80, z-score = -2.12, R) >droSec1.super_13171 636 102 - 1078 AUCCGUCUGCAGCAUGUUGUAGUGCUCCUCUUUGGCACAUAGCGACUCCUUGAGCACCGCUAUGUGCCGCUGGUAGUCCUGACGGGAAUUA----CUUUACUGUGC-------- .((((((.(((((((......))))).......(((((((((((.(((...)))...))))))))))))).((....)).)))))).....----...........-------- ( -36.90, z-score = -2.42, R) >droYak2.chr2L 18061881 102 - 22324452 GUCCGUCUGCAGCAUGUUGUAGUGCUCCUCUUUGGCACAUAGCGACUCCUUGAGCACCGCUAUGUGCCGCUGGUAGUCCUGACGGGAAUUA----CUUUACUGUGC-------- .((((((.(((((((......))))).......(((((((((((.(((...)))...))))))))))))).((....)).)))))).....----...........-------- ( -36.80, z-score = -2.12, R) >droEre2.scaffold_4929 17694996 102 + 26641161 AUCGGUCUGCAGCAUGUUGUAGUGCUCCUCUUUGGCACAUAGCGACUCCUUGAGCACCGCUAUGUGCCGCUGGUAGUCCUGACGGGAAUUA----CUUUACUGUGC-------- ...((.(((((((((......)))))...(..((((((((((((.(((...)))...))))))))))))..))))).))..((((......----.....))))..-------- ( -36.00, z-score = -1.75, R) >droAna3.scaffold_13266 19673573 102 + 19884421 GUCCGUCUGCAGCAUGUUGUAGUGCUCCUCUUUGGCACAUAGCGACUCCUUGAGCACCGCUAUGUGCCGCUGGUAGUCCUAAUGGGAAUUA----CUUUACUGUGC-------- ........(((((((......))))).......(((((((((((.(((...)))...))))))))))))).((((((((.....)).))))----)).........-------- ( -33.00, z-score = -1.38, R) >dp4.chr3 13637826 102 - 19779522 GUCCGAAUGCAGCAUGUUGUAGUGCUCCUCUUUGGCACAUAGCGACUCCUUGAGCACCGCUAUGUGCCGCUGAUAGUCCUAGCGGGAAUUA----CGUUACUGUGC-------- ........((((.....((((((..(((.....(((((((((((.(((...)))...)))))))))))((((.......))))))).))))----))...))))..-------- ( -34.60, z-score = -1.56, R) >droWil1.scaffold_180700 3263430 108 - 6630534 AUCCGUUUGCAGCAUGUUGUAGUGCUCUUCUUUGGCGCAUAGCGAUUCCUUGAGCACCGCUAUAUGCCGCUGAUAGUCCUAAUGAGAGUUA--UUUUUUACUGUUUCUGC---- ........((.((((((.(..((((((.....(.((.....)).)......))))))..).)))))).)).((((((..((((....))))--......)))))).....---- ( -28.20, z-score = -1.20, R) >droVir3.scaffold_10324 191356 104 - 1288806 GUCCGUCUGCAGCAUAUUGUAGUGCUCCUCUUUGGCACAUAGCGACUCCUUGAGCACCGCUAUGUGCCGCUGAUAGUCCUAACGAAAGUUA--UUUUUUACUGUGC-------- (.(..((.(((((((......))))).......(((((((((((.(((...)))...))))))))))))).))..).).((((....))))--.............-------- ( -34.20, z-score = -3.39, R) >droMoj3.scaffold_6496 2009663 103 - 26866924 GUCCGUCUGCAGCAUGUUGUAGUGCUCCUCUUUGGCACAUAGCGAUUCCUUAAGUACCGCUAUGUGCCGCUGAUAGUCCUAAUUUCAGGU---UUGUUCAUUGUGA-------- ....(((.(((((((......))))).......(((((((((((.............))))))))))))).)))...(((......))).---.............-------- ( -29.82, z-score = -2.38, R) >droGri2.scaffold_15245 2889180 104 + 18325388 AUCCGUCUGCAGCAUAUUGUAGUGCUCCUCUUUGGCACAUAGCGACUCCUUGAGCACCGCUAUGUGCCGCUGAUAGUCCUAACGAAAGUUA--UAUUUUACUGUGC-------- ....(((.(((((((......))))).......(((((((((((.(((...)))...))))))))))))).))).....((((....))))--.............-------- ( -34.10, z-score = -3.45, R) >triCas2.ChLG7 15375209 85 - 17478683 AUCGGUUUGGAGCAUGUUGUAGUGUUCCUCUUUAGCGCAUAGCGACUCCUUGAGGACAGCGAUAUGGCGCUGGUAGUCUUGGUGU----------------------------- (((((...(((((((......)))))))...((((((((((.((..(((....)))...)).))).))))))).....)))))..----------------------------- ( -24.50, z-score = 0.02, R) >anoGam1.chr3R 52792833 114 - 53272125 AUCUGAUUGCAACAUAUUGUAAUGUUCUUCUUUGGCGCAUAGUGAUUCUUUCAGAACGGCAAUAUGACGUUGGUAAUCCUAGAAGGAUACAAAUGUAUUAGUAUGCUUUCGAUA (((.((..(((.((((((((...(((((.......(((...)))........))))).))))))))(((((.(((.(((.....)))))).))))).......)))..))))). ( -27.26, z-score = -1.33, R) >apiMel3.Group9 6389995 85 + 10282195 GUCAGCCUGCAGCAUGUUGUAGUGUUCCUCCUUGGCGGCGAGCGACUCUUUGAGCACGGUGAUGUGCCGCUGAUAAUCCUGGUGC----------------------------- ((((((.....((((((..(.((((((.....(.((.....)).)......)))))).)..)))))).))))))...........----------------------------- ( -24.00, z-score = 1.17, R) >consensus GUCCGUCUGCAGCAUGUUGUAGUGCUCCUCUUUGGCACAUAGCGACUCCUUGAGCACCGCUAUGUGCCGCUGGUAGUCCUAACGGGAAUUA____CUUUACUGUGC________ ........((.((((((.(..((((((.....(.((.....)).)......))))))..).)))))).))............................................ (-18.78 = -18.59 + -0.18)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:10:25 2011