
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 5,333,711 – 5,333,803 |
| Length | 92 |
| Max. P | 0.924536 |

| Location | 5,333,711 – 5,333,803 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 83.93 |
| Shannon entropy | 0.27595 |
| G+C content | 0.48900 |
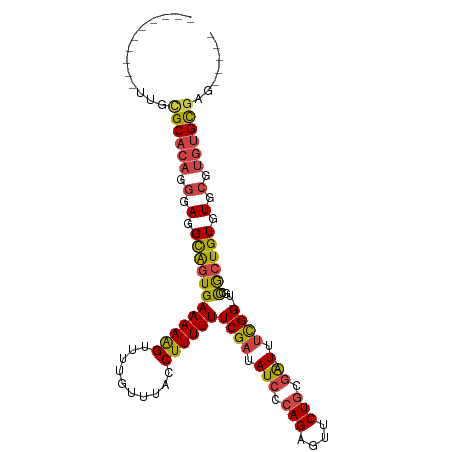
| Mean single sequence MFE | -32.75 |
| Consensus MFE | -21.68 |
| Energy contribution | -21.88 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.924536 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 5333711 92 + 21146708 ----------UUGCGCACAGGGAGGCAGUGAAAAAGUUUUGUUUACCUUUUUUCGAUAUCCCAGAGUUCUGCGA--UUUCGG--UGUCGCUGUGUGCGUGUGCGAG----- ----------...((((((.(.(.((((((((((((..........))))).((((.(((.(((....))).))--).))))--..))))))).).).))))))..----- ( -32.80, z-score = -2.75, R) >droAna3.scaffold_13266 19594933 111 - 19884421 UAGCACGCAAAAUAGCAGUCGGAGGUGGUGAAAAAGUUUUGUUUACCUUUUUUCGAUAUCCGAGAAUUCUGCUGCGUUCUGGCCUGCUGGUGUGUGAGUGUGUGCCUUCGG ..(((((((..(((((((.(((((..((((((.........))))))..((((((.....)))))))))))))))......(((....))).)))...)))))))...... ( -33.50, z-score = -1.33, R) >droSim1.chr2R 3984316 92 + 19596830 ----------UUGCGCACAGGGAGGCAGUGAAAAAGUUUUGUUUACCUUUUUUCGAUAUCCCAGAGUUCUGCGA--UUUCGG--UGUCGCUGUGUGCGUGUGCGAG----- ----------...((((((.(.(.((((((((((((..........))))).((((.(((.(((....))).))--).))))--..))))))).).).))))))..----- ( -32.80, z-score = -2.75, R) >droSec1.super_1 2959014 92 + 14215200 ----------UAGCGCACAGGGAGGCAGUGAAAAAGUUUUGUUUACCUUUUUUCGAUAUCCCAGAGUUCUGCGA--UUUCGG--UGUCGCUGUGUGCGUGUGCGAG----- ----------...((((((.(.(.((((((((((((..........))))).((((.(((.(((....))).))--).))))--..))))))).).).))))))..----- ( -32.80, z-score = -2.83, R) >droYak2.chr2L 17989745 92 + 22324452 ----------UUGUGCACAGGGAGGCAGUGAAAAGGUUUUGUUUACCUUUUUUCGAUAUCCCAGAGUUCUGCGA--UUUCGG--UGCCACUGUGUGCGUGUGCGAG----- ----------...((((((.(.(.(((((((((((((.......))))))).((((.(((.(((....))).))--).))))--...)))))).).).))))))..----- ( -35.90, z-score = -3.82, R) >droEre2.scaffold_4929 17623571 92 - 26641161 ----------UGGCGCUCAGGGAGGCAGUGAAAAAGUUUUGUUUACCUUUUUUCGAUAUCCCAGAGUUCUGCGA--UUUCGG--UGCCACUGUGUGCGUGUGCGAG----- ----------...(((.((.(.(.((((((((((((..........))))))((((.(((.(((....))).))--).))))--...)))))).).).)).)))..----- ( -28.70, z-score = -1.19, R) >consensus __________UUGCGCACAGGGAGGCAGUGAAAAAGUUUUGUUUACCUUUUUUCGAUAUCCCAGAGUUCUGCGA__UUUCGG__UGCCGCUGUGUGCGUGUGCGAG_____ .............((((((.(.(.((((((((((((..........)))))).(((..((.(((....))).))....)))......)))))).).).))))))....... (-21.68 = -21.88 + 0.20)
| Location | 5,333,711 – 5,333,803 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 83.93 |
| Shannon entropy | 0.27595 |
| G+C content | 0.48900 |
| Mean single sequence MFE | -24.96 |
| Consensus MFE | -14.47 |
| Energy contribution | -14.77 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.910309 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 5333711 92 - 21146708 -----CUCGCACACGCACACAGCGACA--CCGAAA--UCGCAGAACUCUGGGAUAUCGAAAAAAGGUAAACAAAACUUUUUCACUGCCUCCCUGUGCGCAA---------- -----.........((.(((((.((..--.(((.(--((.(((....))).))).)))..(((((((.......))))))).......)).))))).))..---------- ( -24.20, z-score = -2.76, R) >droAna3.scaffold_13266 19594933 111 + 19884421 CCGAAGGCACACACUCACACACCAGCAGGCCAGAACGCAGCAGAAUUCUCGGAUAUCGAAAAAAGGUAAACAAAACUUUUUCACCACCUCCGACUGCUAUUUUGCGUGCUA .....(((((..................((......)).(((((((..(((((.......(((((((.......))))))).......))))).....)))))))))))). ( -23.64, z-score = -1.26, R) >droSim1.chr2R 3984316 92 - 19596830 -----CUCGCACACGCACACAGCGACA--CCGAAA--UCGCAGAACUCUGGGAUAUCGAAAAAAGGUAAACAAAACUUUUUCACUGCCUCCCUGUGCGCAA---------- -----.........((.(((((.((..--.(((.(--((.(((....))).))).)))..(((((((.......))))))).......)).))))).))..---------- ( -24.20, z-score = -2.76, R) >droSec1.super_1 2959014 92 - 14215200 -----CUCGCACACGCACACAGCGACA--CCGAAA--UCGCAGAACUCUGGGAUAUCGAAAAAAGGUAAACAAAACUUUUUCACUGCCUCCCUGUGCGCUA---------- -----..(((((((((.....)))...--.(((.(--((.(((....))).))).))).....(((((................)))))...))))))...---------- ( -23.79, z-score = -2.68, R) >droYak2.chr2L 17989745 92 - 22324452 -----CUCGCACACGCACACAGUGGCA--CCGAAA--UCGCAGAACUCUGGGAUAUCGAAAAAAGGUAAACAAAACCUUUUCACUGCCUCCCUGUGCACAA---------- -----.........(((((..(.((((--.(((.(--((.(((....))).))).)))..(((((((.......)))))))...)))).)..)))))....---------- ( -30.00, z-score = -4.61, R) >droEre2.scaffold_4929 17623571 92 + 26641161 -----CUCGCACACGCACACAGUGGCA--CCGAAA--UCGCAGAACUCUGGGAUAUCGAAAAAAGGUAAACAAAACUUUUUCACUGCCUCCCUGAGCGCCA---------- -----........(((.((..(.((((--.(((.(--((.(((....))).))).)))..(((((((.......)))))))...)))).)..)).)))...---------- ( -23.90, z-score = -2.02, R) >consensus _____CUCGCACACGCACACAGCGACA__CCGAAA__UCGCAGAACUCUGGGAUAUCGAAAAAAGGUAAACAAAACUUUUUCACUGCCUCCCUGUGCGCAA__________ .............((((((..((((............))))........((((.......(((((((.......))))))).......))))))))))............. (-14.47 = -14.77 + 0.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:10:22 2011