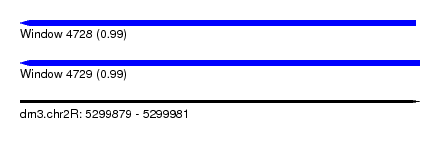
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 5,299,879 – 5,299,981 |
| Length | 102 |
| Max. P | 0.993834 |

| Location | 5,299,879 – 5,299,980 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 75.21 |
| Shannon entropy | 0.40986 |
| G+C content | 0.34813 |
| Mean single sequence MFE | -25.42 |
| Consensus MFE | -12.90 |
| Energy contribution | -15.82 |
| Covariance contribution | 2.92 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.11 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.65 |
| SVM RNA-class probability | 0.993834 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 5299879 101 - 21146708 GUGCGAAUUGCUUGCAUUUGGACAGCUUUUCCAAACAAAUAUUUCUUACGACUUUAGGAUAUAUGAAUUUGGAAAAGCUCGAAAACGUGAAAACAAUUCCA---------- ((((((.....)))))).((((.((((((((((((...((((.(((((......))))).))))...)))))))))))).......((....))...))))---------- ( -29.40, z-score = -4.61, R) >droEre2.scaffold_4929 9275563 104 + 26641161 GUGCG-AGUUGCUGCACUUGGGCAGA------AAACACAUAUUUUUUACGACUUUGGCAUAUAUGCAUAUGAAAAAGCUCUAUACCUUAAGAGCAGCACCAAAUUCAGAUU ....(-(((((((((.((((((.(((------(((......))))))..((((((..(((((....)))))..)))).)).....)))))).))))))....))))..... ( -23.80, z-score = -1.30, R) >droYak2.chr2L 17947215 104 - 22324452 GUGCG-AAUUGCUGCACUUGGGCAGA------AAACAAAUAUUUCUUACGACUUGAGGAUAUAUGCAUGUGAAAAAGCUCUAUAACUUGAAAACAAUGCCAAAUUUAGAUU (((((-......)))))...((((..------......((((((((((.....)))))).))))...(((....(((........)))....))).))))........... ( -16.70, z-score = 0.05, R) >droSec1.super_1 2921853 101 - 14215200 GUGCGGAAUUGUUGCAUUUGGACAGCUUUUCCAAACAAAUAUUUCCUACGACUUGAGGAUAUAUGCAUUUGGAAAAGUUCUAAAAAUUAUAAACAAUACUG---------- ...(((.((((((...((((((..(((((((((((...((((.((((........)))).))))...))))))))))))))))).......)))))).)))---------- ( -27.90, z-score = -4.49, R) >droSim1.chr2R 3946676 101 - 19596830 GUGCGGAAUUGUUGCAUUUGGAUAGCUUUUCCAAACAAAUAUUUCCUACGACUUGAGGAUAUAUGCAUUUGGAAAAGCUCUAAAACUUAUAAACAAUACUG---------- ...(((.((((((...((((((..(((((((((((...((((.((((........)))).))))...))))))))))))))))).......)))))).)))---------- ( -29.30, z-score = -5.19, R) >consensus GUGCG_AAUUGUUGCAUUUGGACAGCUUUUCCAAACAAAUAUUUCUUACGACUUGAGGAUAUAUGCAUUUGGAAAAGCUCUAAAACUUAAAAACAAUACCA__________ ((((((.....))))))......((((((((((((...((((.((((........)))).))))...))))))))))))................................ (-12.90 = -15.82 + 2.92)

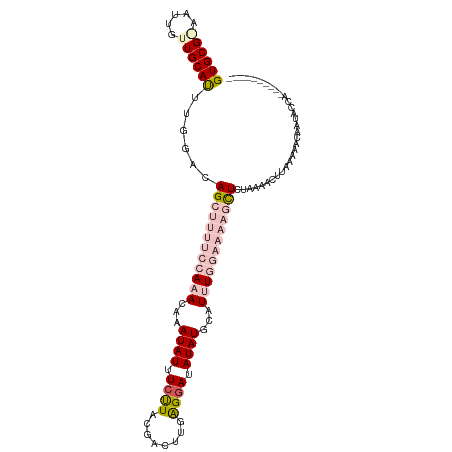
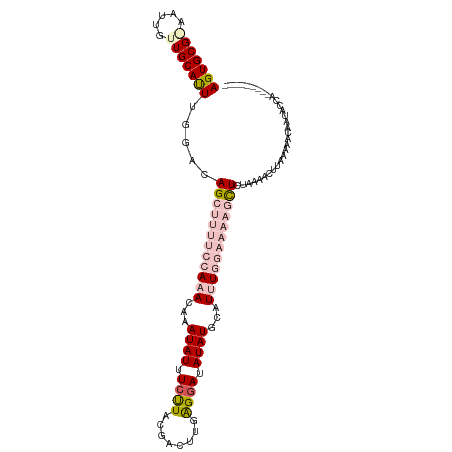
| Location | 5,299,879 – 5,299,981 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 75.84 |
| Shannon entropy | 0.40111 |
| G+C content | 0.34615 |
| Mean single sequence MFE | -25.64 |
| Consensus MFE | -13.46 |
| Energy contribution | -16.38 |
| Covariance contribution | 2.92 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.95 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.39 |
| SVM RNA-class probability | 0.989789 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 5299879 102 - 21146708 AGUGCGAAUUGCUUGCAUUUGGACAGCUUUUCCAAACAAAUAUUUCUUACGACUUUAGGAUAUAUGAAUUUGGAAAAGCUCGAAAACGUGAAAACAAUUCCA--------- (((((((.....)))))))((((.((((((((((((...((((.(((((......))))).))))...)))))))))))).......((....))...))))--------- ( -30.00, z-score = -4.53, R) >droEre2.scaffold_4929 9275564 104 + 26641161 AGUGCG-AGUUGCUGCACUUGGGCAG------AAAACACAUAUUUUUUACGACUUUGGCAUAUAUGCAUAUGAAAAAGCUCUAUACCUUAAGAGCAGCACCAAAUUCAGAU .....(-(((((((((.((((((.((------((((......))))))..((((((..(((((....)))))..)))).)).....)))))).))))))....)))).... ( -23.80, z-score = -1.18, R) >droYak2.chr2L 17947216 104 - 22324452 AGUGCG-AAUUGCUGCACUUGGGCAG------AAAACAAAUAUUUCUUACGACUUGAGGAUAUAUGCAUGUGAAAAAGCUCUAUAACUUGAAAACAAUGCCAAAUUUAGAU ((((((-......))))))..((((.------.......((((((((((.....)))))).))))...(((....(((........)))....))).)))).......... ( -17.20, z-score = 0.01, R) >droSec1.super_1 2921853 102 - 14215200 AGUGCGGAAUUGUUGCAUUUGGACAGCUUUUCCAAACAAAUAUUUCCUACGACUUGAGGAUAUAUGCAUUUGGAAAAGUUCUAAAAAUUAUAAACAAUACUG--------- ....(((.((((((...((((((..(((((((((((...((((.((((........)))).))))...))))))))))))))))).......)))))).)))--------- ( -27.90, z-score = -4.21, R) >droSim1.chr2R 3946676 102 - 19596830 AGUGCGGAAUUGUUGCAUUUGGAUAGCUUUUCCAAACAAAUAUUUCCUACGACUUGAGGAUAUAUGCAUUUGGAAAAGCUCUAAAACUUAUAAACAAUACUG--------- ....(((.((((((...((((((..(((((((((((...((((.((((........)))).))))...))))))))))))))))).......)))))).)))--------- ( -29.30, z-score = -4.82, R) >consensus AGUGCG_AAUUGUUGCAUUUGGACAGCUUUUCCAAACAAAUAUUUCUUACGACUUGAGGAUAUAUGCAUUUGGAAAAGCUCUAAAACUUAAAAACAAUACCA_________ (((((((.....))))))).....((((((((((((...((((.((((........)))).))))...))))))))))))............................... (-13.46 = -16.38 + 2.92)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:10:18 2011