
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 5,296,661 – 5,296,782 |
| Length | 121 |
| Max. P | 0.925755 |

| Location | 5,296,661 – 5,296,752 |
|---|---|
| Length | 91 |
| Sequences | 9 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 59.48 |
| Shannon entropy | 0.81533 |
| G+C content | 0.44220 |
| Mean single sequence MFE | -21.49 |
| Consensus MFE | -9.52 |
| Energy contribution | -9.26 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.50 |
| Mean z-score | -0.92 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.925755 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
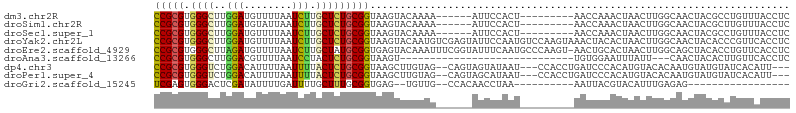
>dm3.chr2R 5296661 91 - 21146708 CCGCGUGGGCUUGGAUGUUUUAAUCUUGCUCUGCGGUAAGUACAAAA------AUUCCACU---------AACCAAACUAACUUGGCAACUACGCCUGUUUACCUC ..((((((...((((..((((...((((((....))))))...))))------..))))..---------..((((......))))...))))))........... ( -21.30, z-score = -1.44, R) >droSim1.chr2R 3943403 91 - 19596830 CCGCGUGGGCUUGGAUGUAUUAAUCUUGCUCUGCGGUAAGUACAAAA------AUUCCACU---------AACCAAACUAACUUGGCAACUACGCUUGUUUACCUC ..((((((...((((.........((((((....)))))).......------..))))..---------..((((......))))...))))))........... ( -20.77, z-score = -1.40, R) >droSec1.super_1 2918726 91 - 14215200 CCGCGUGGGCUUGGAUGUUUUAAUCUUGCUCUGCGGUAAGUACAAAA------AUUCCACU---------AACCAAACUAACUUGGCAACUACGCCUGUUUACCUC ..((((((...((((..((((...((((((....))))))...))))------..))))..---------..((((......))))...))))))........... ( -21.30, z-score = -1.44, R) >droYak2.chr2L 17943931 106 - 22324452 CCGCGUGGGCUUGGAUGUUUUAAUCUUGCUCUGCGGUAAGUACAAUGUCGAGUAUUCCAAUGUCCAAGUAAACUACACUAACUUGGCAACUACACCCGUUCACCUC (((((.((((..((((......)))).)))))))))...(((...((((((((........((........)).......))))))))..)))............. ( -24.16, z-score = -0.51, R) >droEre2.scaffold_4929 9272254 105 + 26641161 CCGCGUGGGCUUAGAUGUUUUAAUCUUGCUAUGCGGUGAGUACAAAUUUCGGUAUUUCAAUGCCCAAGU-AACUGCACUAACUUGGCAGCUACACCUGUUCACCUC ..((((((.(..((((......)))).)))))))((((((..........(((((....)))))...((-(.((((.(......))))).))).....)))))).. ( -30.00, z-score = -1.88, R) >droAna3.scaffold_13266 11596717 74 + 19884421 CCGCGUGGGCUUGGACGUUUUAAUCCUACUCUGCGGUAAGU-----------------------------UGUGGAAUUUAUU---CAACUACACUUGUUCACCUC (((((.(((...(((........)))..))))))))(((((-----------------------------.((((........---...)))))))))........ ( -16.20, z-score = -0.66, R) >dp4.chr3 9347496 98 - 19779522 CCGCGUGGGUCUGGACAUUUUAAUUUUACUCUGCGGUAAGCUUGUAG--CAGUAGUAUAAU---CCACCUGAUCCCACAUGUACACAAUGUAUGUAUCACAUU--- ....(((((((.((.....(((...(((((((((((.....))))))--.)))))..))).---...)).)).)))))((((((.....))))))........--- ( -22.60, z-score = -0.50, R) >droPer1.super_4 4670716 98 - 7162766 CCGCGUGGGUCUGGACAUUUUAAUUUUACUCUGCGGUAAGCUUGUAG--CAGUAGCAUAAU---CCACCUGAUCCCACAUGUACACAAUGUAUGUAUCACAUU--- ....(((((((.((.....(((...(((((((((((.....))))))--.)))))..))).---...)).)).)))))((((((.....))))))........--- ( -21.90, z-score = -0.19, R) >droGri2.scaffold_15245 12140156 75 + 18325388 UCGAGUGGGACUCGAUAUUUUGAUUUUGCUUUGCGGUGAG--UGUUG--CCACAACCUAA----------AAUUACGUACAUUUGAGAG----------------- (((((.....))))).....((((((((..(((.((..(.--..)..--)).)))..)))----------)))))..............----------------- ( -15.20, z-score = -0.29, R) >consensus CCGCGUGGGCUUGGAUGUUUUAAUCUUGCUCUGCGGUAAGUACAAAG______AUUCCAAU_________AACCACACUAACUUGGCAACUACGCCUGUUCACCUC (((((.((((..(((........))).)))))))))...................................................................... ( -9.52 = -9.26 + -0.27)
| Location | 5,296,691 – 5,296,782 |
|---|---|
| Length | 91 |
| Sequences | 11 |
| Columns | 123 |
| Reading direction | reverse |
| Mean pairwise identity | 62.00 |
| Shannon entropy | 0.71736 |
| G+C content | 0.43045 |
| Mean single sequence MFE | -22.13 |
| Consensus MFE | -9.94 |
| Energy contribution | -9.80 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.64 |
| Mean z-score | -0.58 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.542225 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
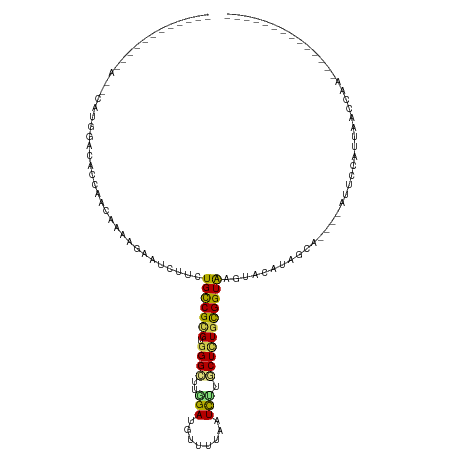
>dm3.chr2R 5296691 91 - 21146708 ------------CACCAUGGAUACCAACAAAAGAAUCCUCUGCCGCGUGGGCUUGGAUGUUUUAAUCUUGCUCUGCGGUAAGUACAAAA------AUUCCACUAACCAA-------------- ------------.....((((..........(((....)))((((((.((((..((((......)))).))))))))))..........------..))))........-------------- ( -22.00, z-score = -1.54, R) >droSim1.chr2R 3943433 91 - 19596830 ------------CGCCAUGGAUACCAACAAAAGAAUCCUCUGCCGCGUGGGCUUGGAUGUAUUAAUCUUGCUCUGCGGUAAGUACAAAA------AUUCCACUAACCAA-------------- ------------.....((((..........(((....)))((((((.((((..((((......)))).))))))))))..........------..))))........-------------- ( -22.00, z-score = -1.30, R) >droSec1.super_1 2918756 91 - 14215200 ------------CGCCAUGGAUACCAACAAAAGAAUCCUCUGCCGCGUGGGCUUGGAUGUUUUAAUCUUGCUCUGCGGUAAGUACAAAA------AUUCCACUAACCAA-------------- ------------.....((((..........(((....)))((((((.((((..((((......)))).))))))))))..........------..))))........-------------- ( -22.00, z-score = -1.23, R) >droYak2.chr2L 17943970 91 - 22324452 ------------------GGAUACCAACAAAAGAAUCCUCUGCCGCGUGGGCUUGGAUGUUUUAAUCUUGCUCUGCGGUAAGUACAAUGUCGAGUAUUCCAAUGUCCAA-------------- ------------------(((((.................(((((((.((((..((((......)))).)))))))))))(((((........)))))....)))))..-------------- ( -26.10, z-score = -2.08, R) >droEre2.scaffold_4929 9272292 91 + 26641161 ------------------GGACGCCAACAAAAGAAUCCUCUGCCGCGUGGGCUUAGAUGUUUUAAUCUUGCUAUGCGGUGAGUACAAAUUUCGGUAUUUCAAUGCCCAA-------------- ------------------...................(((.(((((((((.(..((((......)))).)))))))))))))..........(((((....)))))...-------------- ( -23.40, z-score = -0.78, R) >droAna3.scaffold_13266 11596740 78 + 19884421 ---------------UAUGGACACCAACAAGCGAAUCUUGUGCCGCGUGGGCUUGGACGUUUUAAUCCUACUCUGCGGUAAG-UUGUGGAA----UUU------------------------- ---------------........((((((((.....)))))((((((.(((...(((........)))..)))))))))...-...)))..----...------------------------- ( -19.50, z-score = -0.16, R) >dp4.chr3 9347531 100 - 19779522 -----AACGCGAAAACAUGGACACAAAUAAGAGAAUCUUGUGCCGCGUGGGUCUGGACAUUUUAAUUUUACUCUGCGGUAAGCUUGUAGCA----GUAGUAUAAUCCAC-------------- -----.(((((......((....)).(((((.....)))))..))))).....((((....(((...(((((((((((.....)))))).)----))))..))))))).-------------- ( -19.10, z-score = 0.85, R) >droPer1.super_4 4670751 100 - 7162766 -----AACGCGAAAACAUGGACACAAAUAAGAGAAUCUUGUGCCGCGUGGGUCUGGACAUUUUAAUUUUACUCUGCGGUAAGCUUGUAGCA----GUAGCAUAAUCCAC-------------- -----............((((.(((((((((.....)))))((((((.((((..(((........))).))))))))))....)))).((.----...))....)))).-------------- ( -19.90, z-score = 0.74, R) >droVir3.scaffold_12823 471604 117 - 2474545 --AACAAAACAAAGCUAUGGACACAAACAAGCGCAUUUUGUGCCGAGUGGGACUCGAUGUUUUGAUUUUACUCUGCGGUGAGUG-UUGCCAA---UUGACAUUUGUCACAUUCGUUUCUCCAA --...............((((...((((..((((.....)))).(((((.(((..(((((((((.(..(((((......)))))-..).)))---..)))))).))).))))))))).)))). ( -30.00, z-score = -0.60, R) >droMoj3.scaffold_6496 289040 112 - 26866924 --AAAACAACAAAAAUAUGGACACAAACAAACGAAUCUUAUGCCGUGUAGGCCUCGAUAUUUUGAUUCUGCUCUGUGGUAAGUGUUUGCCUC---UUGUAAUUUCGCAGACGAUAAA------ --................................(((...(((((.(((((..((((....)))).)))))....)))))...((((((...---..........)))))))))...------ ( -15.82, z-score = 1.55, R) >droGri2.scaffold_15245 12140175 96 + 18325388 AAAACAUAACAAA--UAUGGACACAAACAAGCGAAUUUUAUGUCGAGUGGGACUCGAUAUUUUGAUUUUGCUUUGCGGUGAGUG-UUGCCA-----CAACCUAA------------------- .............--...((.(((....(((((((.((.((((((((.....))))))))...)).)))))))....))).(((-....))-----)..))...------------------- ( -23.60, z-score = -1.88, R) >consensus ____________A__CAUGGACACCAACAAAAGAAUCUUCUGCCGCGUGGGCUUGGAUGUUUUAAUCUUGCUCUGCGGUAAGUACAUAGCA____AUUCCAUUAACCAA______________ ........................................(((((((.((((..(((........))).)))))))))))........................................... ( -9.94 = -9.80 + -0.14)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:10:16 2011