| Sequence ID | dm3.chr2R |
|---|---|
| Location | 5,286,139 – 5,286,243 |
| Length | 104 |
| Max. P | 0.996879 |

| Location | 5,286,139 – 5,286,243 |
|---|---|
| Length | 104 |
| Sequences | 12 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 74.10 |
| Shannon entropy | 0.56524 |
| G+C content | 0.41225 |
| Mean single sequence MFE | -26.67 |
| Consensus MFE | -10.70 |
| Energy contribution | -10.95 |
| Covariance contribution | 0.26 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.968818 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
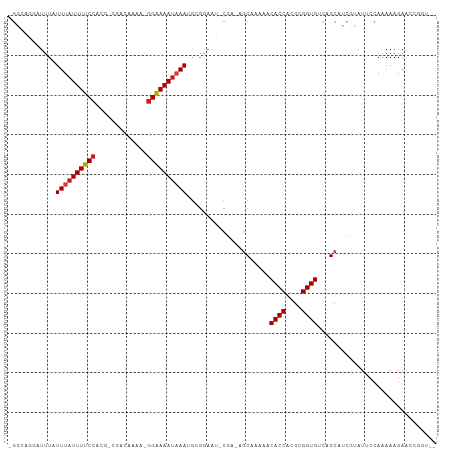
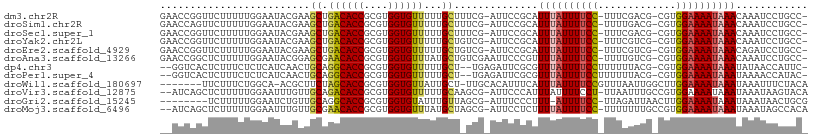
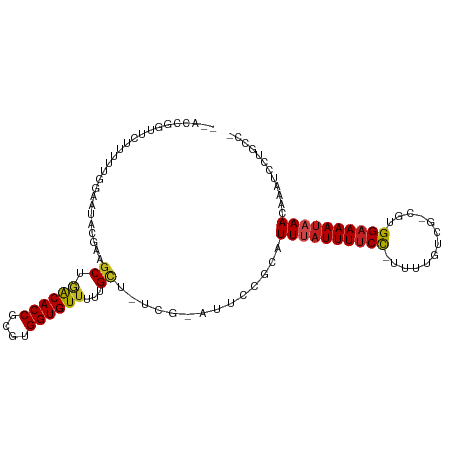
>dm3.chr2R 5286139 104 + 21146708 -GGCAGGAUUUGUUUAUUUUCCACG-CGUCGAAA-GGAAAAUAAAUGCGGAAU-CGAAAGCAAAAACACCACGCGGUGUCAGCUUCGUAUUCCAAAAAGAACCGGUUC -(((.((....(((((((((((...-........-)))))))))))..(((((-((((.((....(((((....)))))..)))))).)))))........)).))). ( -29.50, z-score = -2.25, R) >droSim1.chr2R 3933012 104 + 19596830 -GGCAGGAUUUGUUUAUUUUCCACG-CGUCAAAA-GGAAAAUAAAUGCGGAAU-CGAAAGCAAAAACACCACGCGGUGUCAGCUUCGUAUUCCAAAAAGAACUGGUUC -..(((..((((((((((((((...-........-)))))))))))..(((((-((((.((....(((((....)))))..)))))).)))))...)))..))).... ( -30.00, z-score = -2.95, R) >droSec1.super_1 2908354 104 + 14215200 -GGCAGGAUUUGUUUAUUUUCCACG-CGUCGAAA-GGAAAAUAAAUGCGGAAU-CGAAAGCAAAAACACCACGCGGUGUCAGCUUCGUAUUCCAAAAAGAACCGGUUC -(((.((....(((((((((((...-........-)))))))))))..(((((-((((.((....(((((....)))))..)))))).)))))........)).))). ( -29.50, z-score = -2.25, R) >droYak2.chr2L 17933323 104 + 22324452 -GGCAGGAUUUGUUUAUUUUCCACG-CGACGAAA-GGAAAAUAAAUGCGGAAU-CGACAGCAAAAACACCACGCGGUGUCAGCUUCGUAUUCCAAAAAGAACCGGUUC -(((.((....(((((((((((...-........-)))))))))))..(((((-(((.(((....(((((....)))))..)))))).)))))........)).))). ( -29.50, z-score = -2.25, R) >droEre2.scaffold_4929 9261868 104 - 26641161 -GGCAGGAUCUGUUUAUUUUCCACG-CGACGAAA-GGAAAAUAAAUGCGGAAU-CGACAGCAAAAACACCACGCGGUGUCAGCUUCGUAUUCCAAAAAGAACCGGUUC -(((.((.((((((((((((((...-........-)))))))))))..(((((-(((.(((....(((((....)))))..)))))).)))))....))).)).))). ( -31.80, z-score = -2.96, R) >droAna3.scaffold_13266 11587173 105 - 19884421 -GGCAGGAUUUGUUUAUUUUCCACG-CGACAAAA-GGAAAAUAAAACGGGAAUUCGACAGCAUAAACACCACGCGGUGUUCGCUCCGUAUUCCAAAAAGAGCCGGUUC -(((........((((((((((...-........-))))))))))...(((((.((..(((...((((((....)))))).))).)).))))).......)))..... ( -30.10, z-score = -2.32, R) >dp4.chr3 9336829 102 + 19779522 -GAAUGGUUAUAUUUAUUUUCCACG-CGUAAAAAAGGAAAAUAAACGCGAAUCUCA--AGCAAAAACACCACGCGGUGCCUGCAGUUGAUGAGAGAAAGAGUGACC-- -....((((((.((((((((((...-.........))))))))))......(((((--..(((...((((....)))).......))).)))))......))))))-- ( -22.40, z-score = -1.44, R) >droPer1.super_4 4660052 102 + 7162766 -GUAUGGUUUUAUUUAUUUUCCACG-CGUAAAAAAGGAAAAUAAACGCGAAUCUCA--AGCAAAAACACCACGCGGUGCCUGCAGUUGAUGAGAGAAAGAGUGACC-- -....(((....((((((((((...-.........))))))))))(((...(((((--..(((...((((....)))).......))).)))))......))))))-- ( -20.70, z-score = -0.45, R) >droWil1.scaffold_180697 3436964 99 - 4168966 UGUAGAAAUUUAUUUAUUUUCCAAGCCAAUUAAACGGAAAAUAAAUGAAAUGUGCAA-AGCAAUAACACCACGCGGUGCUAGAAGCGU-UGCCAGAAAGAA------- ........((((((((((((((.............)))))))))))))).((.((((-.((..((.((((....)))).))...)).)-))))).......------- ( -24.92, z-score = -3.45, R) >droVir3.scaffold_12875 1325415 104 + 20611582 UGUACUUAUUUAUUUAUUUUCCACGGCAAAUUAA-AGGAAAAUAAAUGGGAAU-CGCUUGCAAAAACACCACGCGGUGUCUGCAACAAAUUCCAAAAAGAGCUGAU-- .((.(((...((((((((((((............-.))))))))))))(((((-...(((((...(((((....))))).)))))...)))))...))).))....-- ( -27.52, z-score = -3.49, R) >droGri2.scaffold_15245 8657435 97 - 18325388 CGCAGUUAUUUAUUUAUUUUCCAAGUUAAUCUAA-GGAAAAU-AAAGGGAAAU-CGCUAACAAAUACACCACGCGGUGCCUGCAACAGAUUCCAAAAAGA-------- .((((...((..((((((((((............-)))))))-)))..))...-............((((....)))).)))).................-------- ( -19.10, z-score = -1.76, R) >droMoj3.scaffold_6496 4412381 104 + 26866924 UGUGGCUAUUUAUUUAUUUUCCACGGCAAAAAAA-GGAAAAUAAAAGAGGAAU-CGCUAGCAUAAACACCACGCGGUGUUCGCAACAAAUUCCAAAAAGAGCUGAU-- ...((((.(((.((((((((((............-))))))))))...(((((-.....((...((((((....)))))).)).....)))))..))).))))...-- ( -25.00, z-score = -2.45, R) >consensus _GGCAGGAUUUAUUUAUUUUCCACG_CGACAAAA_GGAAAAUAAAUGCGGAAU_CGA_AGCAAAAACACCACGCGGUGUCAGCAUCGUAUUCCAAAAAGAACCGGU__ ............((((((((((.............)))))))))).....................((((....)))).............................. (-10.70 = -10.95 + 0.26)
| Location | 5,286,139 – 5,286,243 |
|---|---|
| Length | 104 |
| Sequences | 12 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 74.10 |
| Shannon entropy | 0.56524 |
| G+C content | 0.41225 |
| Mean single sequence MFE | -28.05 |
| Consensus MFE | -13.43 |
| Energy contribution | -13.40 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.00 |
| SVM RNA-class probability | 0.996879 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
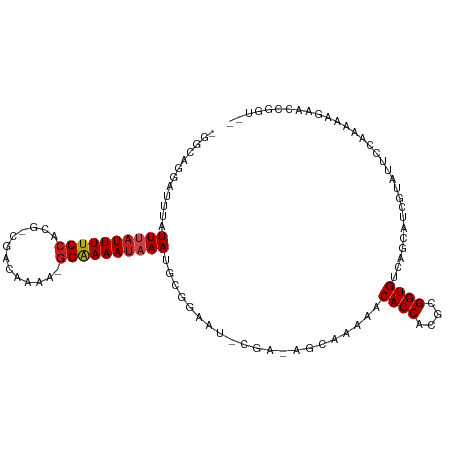
>dm3.chr2R 5286139 104 - 21146708 GAACCGGUUCUUUUUGGAAUACGAAGCUGACACCGCGUGGUGUUUUUGCUUUCG-AUUCCGCAUUUAUUUUCC-UUUCGACG-CGUGGAAAAUAAACAAAUCCUGCC- .....(((....((((((((.((((((.((((((....))))))...)).))))-)))))...((((((((((-........-...)))))))))).)))....)))- ( -29.80, z-score = -2.44, R) >droSim1.chr2R 3933012 104 - 19596830 GAACCAGUUCUUUUUGGAAUACGAAGCUGACACCGCGUGGUGUUUUUGCUUUCG-AUUCCGCAUUUAUUUUCC-UUUUGACG-CGUGGAAAAUAAACAAAUCCUGCC- ....(((........(((((.((((((.((((((....))))))...)).))))-)))))...((((((((((-........-...))))))))))......)))..- ( -29.50, z-score = -2.98, R) >droSec1.super_1 2908354 104 - 14215200 GAACCGGUUCUUUUUGGAAUACGAAGCUGACACCGCGUGGUGUUUUUGCUUUCG-AUUCCGCAUUUAUUUUCC-UUUCGACG-CGUGGAAAAUAAACAAAUCCUGCC- .....(((....((((((((.((((((.((((((....))))))...)).))))-)))))...((((((((((-........-...)))))))))).)))....)))- ( -29.80, z-score = -2.44, R) >droYak2.chr2L 17933323 104 - 22324452 GAACCGGUUCUUUUUGGAAUACGAAGCUGACACCGCGUGGUGUUUUUGCUGUCG-AUUCCGCAUUUAUUUUCC-UUUCGUCG-CGUGGAAAAUAAACAAAUCCUGCC- .....(((....((((((((.((((((.((((((....))))))...))).)))-)))))...((((((((((-........-...)))))))))).)))....)))- ( -29.80, z-score = -2.16, R) >droEre2.scaffold_4929 9261868 104 + 26641161 GAACCGGUUCUUUUUGGAAUACGAAGCUGACACCGCGUGGUGUUUUUGCUGUCG-AUUCCGCAUUUAUUUUCC-UUUCGUCG-CGUGGAAAAUAAACAGAUCCUGCC- .....((.(((....(((((.((((((.((((((....))))))...))).)))-)))))...((((((((((-........-...)))))))))).))).))....- ( -31.90, z-score = -2.56, R) >droAna3.scaffold_13266 11587173 105 + 19884421 GAACCGGCUCUUUUUGGAAUACGGAGCGAACACCGCGUGGUGUUUAUGCUGUCGAAUUCCCGUUUUAUUUUCC-UUUUGUCG-CGUGGAAAAUAAACAAAUCCUGCC- .....(((.......(((((.(((((((((((((....)))))))..))).))).))))).((((.(((((((-........-...))))))))))).......)))- ( -30.50, z-score = -2.06, R) >dp4.chr3 9336829 102 - 19779522 --GGUCACUCUUUCUCUCAUCAACUGCAGGCACCGCGUGGUGUUUUUGCU--UGAGAUUCGCGUUUAUUUUCCUUUUUUACG-CGUGGAAAAUAAAUAUAACCAUUC- --(((.........(((((......(((((((((....)))))..)))).--))))).....(((((((((((.........-...)))))))))))...)))....- ( -25.40, z-score = -2.61, R) >droPer1.super_4 4660052 102 - 7162766 --GGUCACUCUUUCUCUCAUCAACUGCAGGCACCGCGUGGUGUUUUUGCU--UGAGAUUCGCGUUUAUUUUCCUUUUUUACG-CGUGGAAAAUAAAUAAAACCAUAC- --(((.........(((((......(((((((((....)))))..)))).--))))).....(((((((((((.........-...)))))))))))...)))....- ( -25.40, z-score = -2.41, R) >droWil1.scaffold_180697 3436964 99 + 4168966 -------UUCUUUCUGGCA-ACGCUUCUAGCACCGCGUGGUGUUAUUGCU-UUGCACAUUUCAUUUAUUUUCCGUUUAAUUGGCUUGGAAAAUAAAUAAAUUUCUACA -------.......(((((-(.((...(((((((....)))))))..)).-)))).))....((((((((((((...........))))))))))))........... ( -25.80, z-score = -3.75, R) >droVir3.scaffold_12875 1325415 104 - 20611582 --AUCAGCUCUUUUUGGAAUUUGUUGCAGACACCGCGUGGUGUUUUUGCAAGCG-AUUCCCAUUUAUUUUCCU-UUAAUUUGCCGUGGAAAAUAAAUAAAUAAGUACA --....(((..(((.((((((..(((((((((((....))))))..)))))..)-))))).(((((((((((.-............))))))))))))))..)))... ( -28.72, z-score = -3.28, R) >droGri2.scaffold_15245 8657435 97 + 18325388 --------UCUUUUUGGAAUCUGUUGCAGGCACCGCGUGGUGUAUUUGUUAGCG-AUUUCCCUUU-AUUUUCC-UUAGAUUAACUUGGAAAAUAAAUAAAUAACUGCG --------.......((((..(((((((.(((((....)))))...)).)))))-..)))).(((-(((((((-..((.....)).))))))))))............ ( -23.20, z-score = -2.17, R) >droMoj3.scaffold_6496 4412381 104 - 26866924 --AUCAGCUCUUUUUGGAAUUUGUUGCGAACACCGCGUGGUGUUUAUGCUAGCG-AUUCCUCUUUUAUUUUCC-UUUUUUUGCCGUGGAAAAUAAAUAAAUAGCCACA --....(((.(((..((((((.((((((((((((....)))))))..)).))))-)))))...((((((((((-............)))))))))).))).))).... ( -26.80, z-score = -2.72, R) >consensus __ACCGGUUCUUUUUGGAAUACGAAGCUGACACCGCGUGGUGUUUUUGCU_UCG_AUUCCGCAUUUAUUUUCC_UUUUGUCG_CGUGGAAAAUAAACAAAUCCUGCC_ .........................((.((((((....))))))...))..............((((((((((.............))))))))))............ (-13.43 = -13.40 + -0.03)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:10:15 2011