
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 5,255,762 – 5,255,855 |
| Length | 93 |
| Max. P | 0.974983 |

| Location | 5,255,762 – 5,255,855 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 64.77 |
| Shannon entropy | 0.62292 |
| G+C content | 0.38588 |
| Mean single sequence MFE | -20.85 |
| Consensus MFE | -9.11 |
| Energy contribution | -9.45 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.62 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.970017 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 5255762 93 + 21146708 -----------------CUUGAAAUACACACCUGUUCGGUAUAUCAAGGUCGCUGAUUCGUUUUUUUUUUUUUUUUGCCUGGCAGUAGUAUUUAUGUCUAUCUAUCUUUU -----------------....((((((....((((..((((.((((.......))))..................))))..))))..))))))................. ( -11.21, z-score = 0.37, R) >droSim1.chr2R 3902652 100 + 19596830 UUUGGAUAUUUAUCCGCCUUGAAAUAUACACCUGUUCGGUGUAUCAAGGUCGCUGAUUCGUUUU----------UUGCUUGGCAGUAGUAUUUAUGUCCAUCUAUCUUUU ..(((((((..((.((((((((....((((((.....))))))))))))).((((....((...----------..))....)))).).))..))))))).......... ( -27.00, z-score = -3.61, R) >droSec1.super_1 2877988 101 + 14215200 UUUGGAUAUUUAUCCGCCUUGAAAUAUACACCUGUUCGGUGUAUCAAGGUCGCUGAUUCGUUUUU---------UUGCUUGGCAGUAGUAUUUAUGUCUAUCUAUCUUUU ..(((((((..((.((((((((....((((((.....))))))))))))).((((....((....---------..))....)))).).))..))))))).......... ( -24.90, z-score = -2.98, R) >droYak2.chr2L 17902367 101 + 22324452 UUUGAGUAUUAAUCCGCCUUGAAAUAUACACCUGCUCGGUGUAUCAAGGUCACUGAUUCGCCUUU---------UUGCUUCGCCAUAGUAUUUAUAUCUAUCUUCAUUCC ..(((((((((....(((((((....((((((.....))))))))))))).........((....---------..)).......)))))))))................ ( -21.00, z-score = -2.59, R) >droEre2.scaffold_4929 9231244 101 - 26641161 UUUGGGUAUUUAUCCGCCUUCAGAUAUGCACCUGCUCGGUGUAUCGAGGUCGUAGUAUGGCUUUU---------UUGCUUCGCAGUGGUAUUUAUAUCUAUCUUCGGUCU ..(((((((..((((((((((.(((..(((((.....)))))))))))).....((..(((....---------..)))..)).)))).))..))))))).......... ( -25.70, z-score = -1.22, R) >droWil1.scaffold_181148 3102419 83 - 5435427 ----------------UUUGGGAAUUCGUGACCGUCUUGCG-ACUAAAAU-GCCGUUGCGUCUUU---------UCCCGCGUUUUUUUUACUUGUAUGUAUCUACCCUUG ----------------...(((((..((..((.((.((...-.....)).-)).))..))....)---------))))..........(((......))).......... ( -15.30, z-score = -1.22, R) >consensus UUUGG_UAUUUAUCCGCCUUGAAAUAUACACCUGUUCGGUGUAUCAAGGUCGCUGAUUCGUUUUU_________UUGCUUGGCAGUAGUAUUUAUAUCUAUCUACCUUUU ...............(((((((....((((((.....)))))))))))))............................................................ ( -9.11 = -9.45 + 0.34)

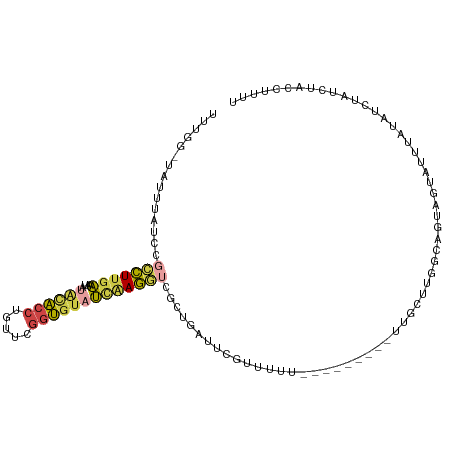
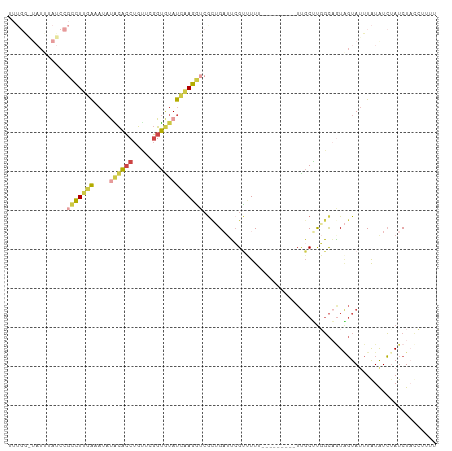
| Location | 5,255,762 – 5,255,855 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 64.77 |
| Shannon entropy | 0.62292 |
| G+C content | 0.38588 |
| Mean single sequence MFE | -17.57 |
| Consensus MFE | -8.07 |
| Energy contribution | -8.35 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.92 |
| SVM RNA-class probability | 0.974983 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 5255762 93 - 21146708 AAAAGAUAGAUAGACAUAAAUACUACUGCCAGGCAAAAAAAAAAAAAAAACGAAUCAGCGACCUUGAUAUACCGAACAGGUGUGUAUUUCAAG----------------- ....(.(((.(((.........)))))).)................................(((((((((((.....))))))....)))))----------------- ( -9.10, z-score = 0.03, R) >droSim1.chr2R 3902652 100 - 19596830 AAAAGAUAGAUGGACAUAAAUACUACUGCCAAGCAA----------AAAACGAAUCAGCGACCUUGAUACACCGAACAGGUGUAUAUUUCAAGGCGGAUAAAUAUCCAAA ..........((((............(((...))).----------............((.((((((((((((.....))))))....))))))))........)))).. ( -21.70, z-score = -3.18, R) >droSec1.super_1 2877988 101 - 14215200 AAAAGAUAGAUAGACAUAAAUACUACUGCCAAGCAA---------AAAAACGAAUCAGCGACCUUGAUACACCGAACAGGUGUAUAUUUCAAGGCGGAUAAAUAUCCAAA ....((((..((..(...........(((...))).---------..............(.((((((((((((.....))))))....))))))))..))..)))).... ( -19.80, z-score = -3.05, R) >droYak2.chr2L 17902367 101 - 22324452 GGAAUGAAGAUAGAUAUAAAUACUAUGGCGAAGCAA---------AAAGGCGAAUCAGUGACCUUGAUACACCGAGCAGGUGUAUAUUUCAAGGCGGAUUAAUACUCAAA ....(((.........................((..---------....)).((((...(.((((((((((((.....))))))....))))))).)))).....))).. ( -19.10, z-score = -0.87, R) >droEre2.scaffold_4929 9231244 101 + 26641161 AGACCGAAGAUAGAUAUAAAUACCACUGCGAAGCAA---------AAAAGCCAUACUACGACCUCGAUACACCGAGCAGGUGCAUAUCUGAAGGCGGAUAAAUACCCAAA ...(((....(((((((...((((........((..---------....))...........((((......))))..)))).)))))))....)))............. ( -18.30, z-score = -1.78, R) >droWil1.scaffold_181148 3102419 83 + 5435427 CAAGGGUAGAUACAUACAAGUAAAAAAAACGCGGGA---------AAAGACGCAACGGC-AUUUUAGU-CGCAAGACGGUCACGAAUUCCCAAA---------------- .....(((......)))...............((((---------(..(((((....))-......((-(....))).))).....)))))...---------------- ( -17.40, z-score = -2.06, R) >consensus AAAAGAUAGAUAGACAUAAAUACUACUGCCAAGCAA_________AAAAACGAAUCAGCGACCUUGAUACACCGAACAGGUGUAUAUUUCAAGGCGGAUAAAUA_CCAAA .............................................................((((((((((((.....))))))....))))))................ ( -8.07 = -8.35 + 0.28)
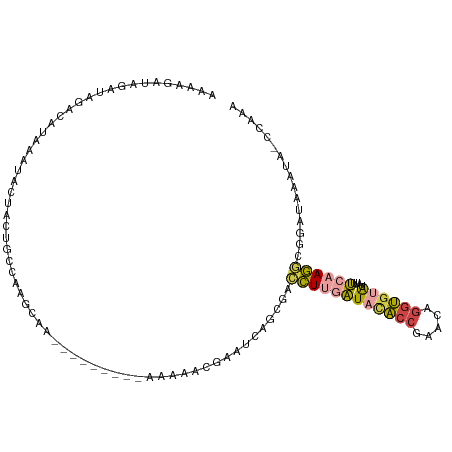
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:10:10 2011