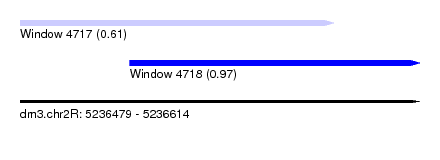
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 5,236,479 – 5,236,614 |
| Length | 135 |
| Max. P | 0.967827 |

| Location | 5,236,479 – 5,236,585 |
|---|---|
| Length | 106 |
| Sequences | 8 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 72.50 |
| Shannon entropy | 0.51751 |
| G+C content | 0.41093 |
| Mean single sequence MFE | -23.55 |
| Consensus MFE | -8.56 |
| Energy contribution | -9.09 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.613531 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 5236479 106 + 21146708 GCAAACAGUAGCGGUGAUUACAAUGUUUUAUUGACUGUUUU--------UACCCACAUAAUUGCCACAGAUUUGGACAUUGUCCAAAUCGACUUGUUUUCCCCUCGA-GACGAGA- ((((...((.(.(((((..(((..(((.....))))))..)--------)))))))....))))....(((((((((...)))))))))..((((((((......))-)))))).- ( -28.50, z-score = -3.41, R) >droSim1.chr2R 3892797 97 + 19596830 GCAAACAGCAGC---------AAUGUUUUAUUGACUGUUUU--------UACCCACAUAAUUGCCACAGAUUUGGACAUUGUCCAAAUCGACUUGUUUUCCCCUCGA-GACGAGA- ((((..((((((---------((((...))))).))))).(--------((......)))))))....(((((((((...)))))))))..((((((((......))-)))))).- ( -24.10, z-score = -3.33, R) >droSec1.super_1 2868211 97 + 14215200 GCAAACAGCAGC---------AAUGUUUUAUUGACUGUUUU--------UACCCACAUAAUUGCCACAGAUUUGGACAUUGUCCAAAUCGACUUGUUUUCCCCUCGA-GACGAGA- ((((..((((((---------((((...))))).))))).(--------((......)))))))....(((((((((...)))))))))..((((((((......))-)))))).- ( -24.10, z-score = -3.33, R) >droYak2.chr2L 17892021 104 + 22324452 GCAAAGAGCAUCGGUGAUUACAAUGUUUUAUUGACUGUUUU--------C-CCCACAUAAUUGCCACAGAUUUGGACAUUGUCCAAAUCGACUUGUUUUCCCCUCGC-GACGAG-- ....((((((((((..(((((((((...)))))..(((...--------.-...)))))))..))...(((((((((...)))))))))))..))))))...((((.-..))))-- ( -23.40, z-score = -1.66, R) >droEre2.scaffold_4929 9220938 106 - 26641161 GCAAACAGCAUCGGUGAUUACAAUGUUUUAUUGACUGUUUU--------UCCCCACAUAAUUGCCACAGUUUUGGACUUUGUCCAAAUCGACUUGUUUUCCCCUCGA-GACGAGC- ((.....)).((((..(((((((((...)))))..(((...--------.....)))))))..))...(.(((((((...))))))).)))((((((((......))-)))))).- ( -21.60, z-score = -1.05, R) >droAna3.scaffold_13266 11552386 99 - 19884421 AUAAACAGCUUCCGUGAUUACUGUAUUUUAUUGACUGUUUU--------U-CUCGCUUAAUUGGCAACAAUUUGGACAAUGUCCAAAUCGUCUUGUUUUCACCCGAA-C------- .............((((..((....................--------.-...(((.....))).((.((((((((...)))))))).))...))..)))).....-.------- ( -16.70, z-score = -0.80, R) >dp4.chr3 9302694 116 + 19779522 AUAAACAGAUUUAGAUAUUAGUCAUUUGGAUCGAGUGUUUUGUGUGCUUUCGCCACAUAAUUGCUAGCGAUUUGGCCAGCUUCCAAAUUGACUUGUUUUUCCUCCACUCACCCGCG .......(((((((((.......)))))))))(((((...((((.((....)))))).((..((.((((((((((.......)))))))).)).))..))....)))))....... ( -26.70, z-score = -1.82, R) >droPer1.super_4 4626083 116 + 7162766 AUAAACAGAUUUGGAUAUUACUCAUUUGGAACGAGUGUUUUGUGUGCUUUCGCCACAUAAUUGCUAGCGAUUUGGCCAGCUUCCAAAUUGACUUGUUUUUCCUCCACUCACCCGCG .....(((((..(.......)..)))))....(((((...((((.((....)))))).((..((.((((((((((.......)))))))).)).))..))....)))))....... ( -23.30, z-score = -0.49, R) >consensus GCAAACAGCAUCGGUGAUUACAAUGUUUUAUUGACUGUUUU________UACCCACAUAAUUGCCACAGAUUUGGACAUUGUCCAAAUCGACUUGUUUUCCCCUCGA_GACGAGA_ ..((((((................(((.....))).................................(((((((((...)))))))))...)))))).................. ( -8.56 = -9.09 + 0.53)
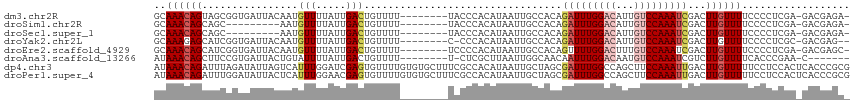
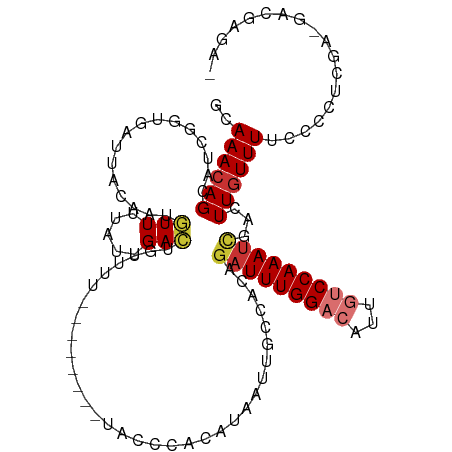
| Location | 5,236,516 – 5,236,614 |
|---|---|
| Length | 98 |
| Sequences | 8 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 67.42 |
| Shannon entropy | 0.60802 |
| G+C content | 0.46333 |
| Mean single sequence MFE | -18.66 |
| Consensus MFE | -7.97 |
| Energy contribution | -8.56 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.967827 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 5236516 98 + 21146708 --------UUUUUACCCACAUAAUUGCCACAGAUUUGGACAUUGUCCAAAUCGACUUGUUUUCCCCUCGAGACGAGACGAGUCUCCCCACUUCCGAAAUCGCUUCU-- --------.................((....(((((((((...)))))))))((((((((((..(.....)..)))))))))).................))....-- ( -22.50, z-score = -3.14, R) >droSim1.chr2R 3892825 96 + 19596830 --------UUUUUACCCACAUAAUUGCCACAGAUUUGGACAUUGUCCAAAUCGACUUGUUUUCCCCUCGAGACGAGACGAGUCUCCC-ACUUCCCAA-UCGCUUCU-- --------.................((....(((((((((...)))))))))((((((((((..(.....)..))))))))))....-.........-..))....-- ( -22.50, z-score = -3.65, R) >droSec1.super_1 2868239 96 + 14215200 --------UUUUUACCCACAUAAUUGCCACAGAUUUGGACAUUGUCCAAAUCGACUUGUUUUCCCCUCGAGACGAGACGAGACUUCC-ACUUCCCAA-UCGCUUCU-- --------.................((....(((((((((...)))))))))..((((((((......))))))))...........-.........-..))....-- ( -19.10, z-score = -2.56, R) >droYak2.chr2L 17892058 84 + 22324452 ---------UUUUCCCCACAUAAUUGCCACAGAUUUGGACAUUGUCCAAAUCGACUUGUUUUCCCCUCGCGACGAGAC-----UCCC----G----AAUCGCUUUC-- ---------................((....(((((((((...)))))))))..((((((..........))))))..-----....----.----....))....-- ( -16.10, z-score = -1.95, R) >droEre2.scaffold_4929 9220975 89 - 26641161 --------UUUUUCCCCACAUAAUUGCCACAGUUUUGGACUUUGUCCAAAUCGACUUGUUUUCCCCUCGAGACGAGCC-----GCCC----GCCCGAAUCGCUUUU-- --------.................((......(((((((...))))))).((.((((((((......)))))))).)-----)...----.........))....-- ( -14.90, z-score = -1.05, R) >droAna3.scaffold_13266 11552423 91 - 19884421 ---------UUUUUCUCGCUUAAUUGGCAACAAUUUGGACAAUGUCCAAAUCGUCUUGUUUUCACC-CGAACCUUCCCGA---CCCC----GUCAGAGCCAUCUAGCA ---------........(((....((((.((.((((((((...)))))))).))............-...........((---(...----)))...))))...))). ( -20.00, z-score = -2.68, R) >dp4.chr3 9302731 99 + 19779522 UUUUGUGUGCUUUCGCCACAUAAUUGCUAGCGAUUUGGCCAGCUUCCAAAUUGACUUGUUUUUCCUCCACUCACCCGCGACCACCCA------CAAAAUGGUUAA--- ...((((.((....)))))).((..((.((((((((((.......)))))))).)).))..))...............(((((....------.....)))))..--- ( -17.10, z-score = -0.36, R) >droPer1.super_4 4626120 99 + 7162766 UUUUGUGUGCUUUCGCCACAUAAUUGCUAGCGAUUUGGCCAGCUUCCAAAUUGACUUGUUUUUCCUCCACUCACCCGCGACCACCCA------CAAAAUGGUUAA--- ...((((.((....)))))).((..((.((((((((((.......)))))))).)).))..))...............(((((....------.....)))))..--- ( -17.10, z-score = -0.36, R) >consensus ________UUUUUCCCCACAUAAUUGCCACAGAUUUGGACAUUGUCCAAAUCGACUUGUUUUCCCCUCGAGACGAGACGA___UCCC____GCCAAAAUCGCUUAU__ ...............................(((((((((...)))))))))........................................................ ( -7.97 = -8.56 + 0.59)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:10:09 2011