| Sequence ID | dm3.chr2R |
|---|---|
| Location | 5,202,691 – 5,202,791 |
| Length | 100 |
| Max. P | 0.546029 |

| Location | 5,202,691 – 5,202,791 |
|---|---|
| Length | 100 |
| Sequences | 7 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 72.58 |
| Shannon entropy | 0.53064 |
| G+C content | 0.39760 |
| Mean single sequence MFE | -25.03 |
| Consensus MFE | -11.36 |
| Energy contribution | -13.00 |
| Covariance contribution | 1.64 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.546029 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 5202691 100 + 21146708 --------UACUCUUUUGUGGCUUAACUGGAGCAUGAUGGAGAUGCAACAUGCAACAGAUUUGCUGUGGUAAUGGAAAGCUUGUUGAAACAACUCAACAACGUCCAUU --------.......(..((((..(.(((..(((((.((......)).)))))..))).)..))))..).((((((....(((((((......)))))))..)))))) ( -28.70, z-score = -2.15, R) >droSim1.chr2R 3862569 100 + 19596830 --------UACUCUUUUGUGGCUUAACUGGAGCAUGAUGGAGAUGCAACAUGCAACAGAUUUGCUGUGGUAAUGGAAAGCUUGUUGAAACAACUCAACAACGUCCAUU --------.......(..((((..(.(((..(((((.((......)).)))))..))).)..))))..).((((((....(((((((......)))))))..)))))) ( -28.70, z-score = -2.15, R) >droSec1.super_1 2834658 100 + 14215200 --------UACUCUUUUGUGGCUUAACUGUAGCAUGAUGGAGAUGCAACAUGCAACAGAUUUGCUGUGGUAAUGGAAAGCUUGUUGAAACAACUCAACAACGUCCAUU --------.......(..((((..(.((((.(((((.((......)).))))).)))).)..))))..).((((((....(((((((......)))))))..)))))) ( -29.50, z-score = -2.39, R) >droYak2.chr2L 17858332 94 + 22324452 --------CGCUCUUUUGUGGUUUAACUGGAGCAUGAUGGAGAUGCAACAUGCAACAGAUUUUCCGUGGUAAUGGAAAGCUUGUUGAAACAACUC------GUCCAUU --------.(((((...((......)).)))))..((((((((.........((((((.((((((((....)))))))).)))))).......))------.)))))) ( -28.09, z-score = -2.09, R) >droEre2.scaffold_4929 9186822 94 - 26641161 --------CCCUCUUUUGUGGCUUAACUGGAGCAUGAUGGAGAUGCAACAUGCAACAGAUUUGCCGUGGUAAUGGAAAGCUUGUUGAAACAACUC------GUCCAGC --------.......(..((((..(.(((..(((((.((......)).)))))..))).)..))))..)...((((.((.((((....)))))).------.)))).. ( -25.40, z-score = -0.71, R) >droAna3.scaffold_12430 35707 102 - 45982 UUUUCUGCCUCGAUUUUGCGGUUUAACUGGAGCAUGAUGAAAAUGCAAUAUGCAACAGAUUUGCACUUCUAAUGGAAAGCUUGUUGAAUCAACUU------GUUCAUU ...........(((((.((((.....(((..(((((.((......)).)))))..)))....((..(((....)))..)))))).))))).....------....... ( -17.90, z-score = 0.87, R) >droMoj3.scaffold_6496 4310123 88 + 26866924 ---------UUUUUUUUUUGGGUGCUCCCGAA----ACGAAAGGGCCAU---UAAGUUAUUCAAAUUGAAAAUAGAAUACUUGACAGCAUAAUUAAUUGGAAAU---- ---------(((((.(((((((....))))))----).))))).((..(---(((((.((((............))))))))))..))................---- ( -16.90, z-score = -1.12, R) >consensus ________CACUCUUUUGUGGCUUAACUGGAGCAUGAUGGAGAUGCAACAUGCAACAGAUUUGCUGUGGUAAUGGAAAGCUUGUUGAAACAACUCA_____GUCCAUU ...............(..((((....(((..(((((.((......)).)))))..)))....))))..).((((((......((((...)))).........)))))) (-11.36 = -13.00 + 1.64)

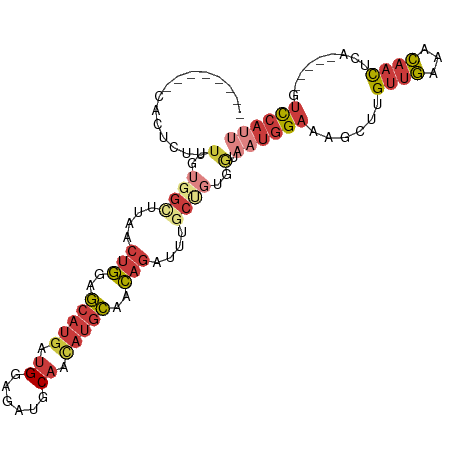

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:10:02 2011