| Sequence ID | dm3.chr2R |
|---|---|
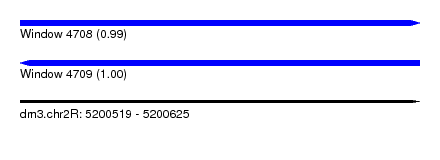
| Location | 5,200,519 – 5,200,625 |
| Length | 106 |
| Max. P | 0.998300 |

| Location | 5,200,519 – 5,200,625 |
|---|---|
| Length | 106 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 88.07 |
| Shannon entropy | 0.25509 |
| G+C content | 0.34109 |
| Mean single sequence MFE | -25.68 |
| Consensus MFE | -22.95 |
| Energy contribution | -22.95 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.35 |
| SVM RNA-class probability | 0.989000 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 5200519 106 + 21146708 GUUGCAUUUUGUUCAAACUGCAGUUGAAGUAAAUGCCUGUUGAUGAAUUCUCGAAUG-CAACUUUCAUCAAUCCAGACUAUUUACUUUAAACACAGUCCAACAUUGU------------ ...(((...((((...((((...((((((((((((.(((((((((((..........-.....))))))))..)))..))))))))))))...))))..)))).)))------------ ( -25.46, z-score = -2.63, R) >droSim1.chr2R 3860394 106 + 19596830 GUUGCAUUUUGUUCAAACUGCAGUUGAAGUAAAUGCCUGUUGAUGAAUUCUCGAAUG-CAACUUUCAUCAAUCCAGACUAUUUACUUUAAACACAGUCCAACAUUGU------------ ...(((...((((...((((...((((((((((((.(((((((((((..........-.....))))))))..)))..))))))))))))...))))..)))).)))------------ ( -25.46, z-score = -2.63, R) >droSec1.super_1 2832520 106 + 14215200 GUUGCAUUUUGUUCAAACUGCAGUUGAAGUAAAUGCCUGUUGAUGAAUUCUCGAAUG-CAACUUUCAUCAAUCCAGACUAUUUACUUUAAACACAGUCCAACAUUGU------------ ...(((...((((...((((...((((((((((((.(((((((((((..........-.....))))))))..)))..))))))))))))...))))..)))).)))------------ ( -25.46, z-score = -2.63, R) >droYak2.chr2L 17856059 106 + 22324452 GUUGCAUUUCGUUCAAACUGCAGUUGAAGUAAAUGCCUGUUGAUGAAUUCUCGAAUG-CAACUUUCAUCAAUCCAGACUAUUUACUUUAAACACAGUCCAACAUUGU------------ ..........(((...((((...((((((((((((.(((((((((((..........-.....))))))))..)))..))))))))))))...))))..))).....------------ ( -23.76, z-score = -2.22, R) >droEre2.scaffold_4929 9184711 106 - 26641161 GUUGCAUUUCGUUCAAACUGCAGUUGAAGUAAAUGCCUGUUGAUGAAUUCUCGAAUG-CAACUUUCAUCAAUCCAGACUAUUUACUUUAAACACAGUCCAACAUUGU------------ ..........(((...((((...((((((((((((.(((((((((((..........-.....))))))))..)))..))))))))))))...))))..))).....------------ ( -23.76, z-score = -2.22, R) >droAna3.scaffold_12430 33151 106 - 45982 GUUGAAUUUCGUUCGAACUGCAGUUGAAGUAAAUGCCUGUUGAUGAAUUCUCGAAUA-CAACUUUCAUCAAUCCAGACUAUUUACUUUAAGCACAGUCCAACAUUGU------------ ((((....((....))((((...((((((((((((.(((((((((((..........-.....))))))))..)))..))))))))))))...)))).)))).....------------ ( -24.16, z-score = -2.24, R) >dp4.chr3 9267907 118 + 19779522 GUUGCAUAUCGUUUAAACUGCAGUUGAAGUAAAUGCCUGUUGAUGAAUUCUUGAAUA-UAACUUUCAUCAAUCCAGACUAUUUACUUUAAGCACAGUCCAACAUUAUUUCUGUGUGUGU .(((((............)))))((((((((((((.(((((((((((..........-.....))))))))..)))..))))))))))))((((((.............)))))).... ( -26.38, z-score = -2.05, R) >droPer1.super_4 4590330 118 + 7162766 GUUGCAUAUCGUUUAAACUGCAGUUGAAGUAAAUGCCUGUUGAUGAAUUCUUGAAUA-UAACUUUCAUCAAUCCAGACUAUUUACUUUAAGCACAGUCCAACAUUAUUUCUGUGUGUGU .(((((............)))))((((((((((((.(((((((((((..........-.....))))))))..)))..))))))))))))((((((.............)))))).... ( -26.38, z-score = -2.05, R) >droWil1.scaffold_180697 3315652 113 - 4168966 GUUGCAUUUUGUUCAAACUGCAGUUGAAGUAAAUGCCUGUUGAUGAAUUCUUGAAUA-UAACUUUCAUCAAUCCAGACUAUUUACUUUAAGCACAGUUCAACAUUAUGAUCUCU----- ....(((..((((..(((((...((((((((((((.(((((((((((..........-.....))))))))..)))..))))))))))))...))))).))))..)))......----- ( -27.36, z-score = -2.95, R) >droVir3.scaffold_12875 1241658 116 + 20611582 GUUGCAUUUCGUUUAAACUGCAGUUGAAGUAAAUGCCUGUUGAUGAAUUCUUGAAUA-AAACUUUCAUCAAUCCAGACUAUUUACUUUAAGCACAGUUCAACA-UUUGUGAGCGACGC- (((((..................((((((((((((.(((((((((((..........-.....))))))))..)))..)))))))))))).(((((.......-.))))).)))))..- ( -29.16, z-score = -2.42, R) >droMoj3.scaffold_6496 4307040 118 + 26866924 GUUGCAUUUCGUUUAAACUGCAGUUGAAGUAAAUGCCUGUUGAUGAAUUCUUAGAUAUAAACUUUCAUCAAUCCAGACUAUUUACUUUAAGCACAGUUCAACAGUUUAUUCGCGAUAU- (((((.....(((..(((((...((((((((((((.(((((((((((...(((....)))...))))))))..)))..))))))))))))...))))).))).(......))))))..- ( -27.20, z-score = -2.78, R) >droGri2.scaffold_15245 8580541 111 - 18325388 GUUGCAUUUCGUUUAAACUGCAGUUGAAGUAAAUGCCUGUUGAUGAAUUCUUGAAUA-AAACUUUCAUCAAUCCAGACUAUUUACUUUAAGCACAGUUCAACAGAUAGAUAU------- ((((...........(((((...((((((((((((.(((((((((((..........-.....))))))))..)))..))))))))))))...)))))))))..........------- ( -23.56, z-score = -1.84, R) >consensus GUUGCAUUUCGUUCAAACUGCAGUUGAAGUAAAUGCCUGUUGAUGAAUUCUCGAAUA_CAACUUUCAUCAAUCCAGACUAUUUACUUUAAGCACAGUCCAACAUUGU____________ ..........(((...((((...((((((((((((.(((((((((((................))))))))..)))..))))))))))))...))))..)))................. (-22.95 = -22.95 + -0.00)

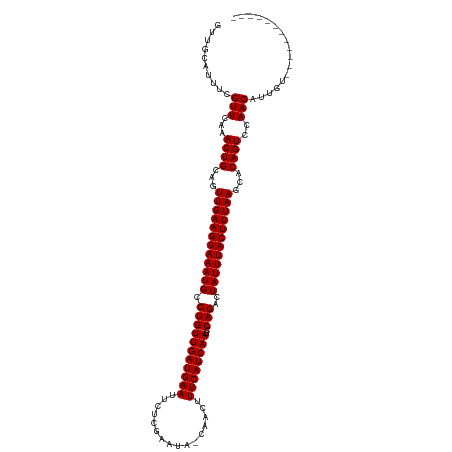
| Location | 5,200,519 – 5,200,625 |
|---|---|
| Length | 106 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 88.07 |
| Shannon entropy | 0.25509 |
| G+C content | 0.34109 |
| Mean single sequence MFE | -30.38 |
| Consensus MFE | -29.95 |
| Energy contribution | -29.43 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.12 |
| Structure conservation index | 0.99 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.31 |
| SVM RNA-class probability | 0.998300 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 5200519 106 - 21146708 ------------ACAAUGUUGGACUGUGUUUAAAGUAAAUAGUCUGGAUUGAUGAAAGUUG-CAUUCGAGAAUUCAUCAACAGGCAUUUACUUCAACUGCAGUUUGAACAAAAUGCAAC ------------....(((..((((((..((.((((((((.(((((...(((((((..((.-......))..))))))).))))))))))))).))..))))))..).))......... ( -30.00, z-score = -2.85, R) >droSim1.chr2R 3860394 106 - 19596830 ------------ACAAUGUUGGACUGUGUUUAAAGUAAAUAGUCUGGAUUGAUGAAAGUUG-CAUUCGAGAAUUCAUCAACAGGCAUUUACUUCAACUGCAGUUUGAACAAAAUGCAAC ------------....(((..((((((..((.((((((((.(((((...(((((((..((.-......))..))))))).))))))))))))).))..))))))..).))......... ( -30.00, z-score = -2.85, R) >droSec1.super_1 2832520 106 - 14215200 ------------ACAAUGUUGGACUGUGUUUAAAGUAAAUAGUCUGGAUUGAUGAAAGUUG-CAUUCGAGAAUUCAUCAACAGGCAUUUACUUCAACUGCAGUUUGAACAAAAUGCAAC ------------....(((..((((((..((.((((((((.(((((...(((((((..((.-......))..))))))).))))))))))))).))..))))))..).))......... ( -30.00, z-score = -2.85, R) >droYak2.chr2L 17856059 106 - 22324452 ------------ACAAUGUUGGACUGUGUUUAAAGUAAAUAGUCUGGAUUGAUGAAAGUUG-CAUUCGAGAAUUCAUCAACAGGCAUUUACUUCAACUGCAGUUUGAACGAAAUGCAAC ------------......(..((((((..((.((((((((.(((((...(((((((..((.-......))..))))))).))))))))))))).))..))))))..)............ ( -29.40, z-score = -2.56, R) >droEre2.scaffold_4929 9184711 106 + 26641161 ------------ACAAUGUUGGACUGUGUUUAAAGUAAAUAGUCUGGAUUGAUGAAAGUUG-CAUUCGAGAAUUCAUCAACAGGCAUUUACUUCAACUGCAGUUUGAACGAAAUGCAAC ------------......(..((((((..((.((((((((.(((((...(((((((..((.-......))..))))))).))))))))))))).))..))))))..)............ ( -29.40, z-score = -2.56, R) >droAna3.scaffold_12430 33151 106 + 45982 ------------ACAAUGUUGGACUGUGCUUAAAGUAAAUAGUCUGGAUUGAUGAAAGUUG-UAUUCGAGAAUUCAUCAACAGGCAUUUACUUCAACUGCAGUUCGAACGAAAUUCAAC ------------....(((((((((((..((.((((((((.(((((...(((((((..(((-....)))...))))))).))))))))))))).))..))))))).))))......... ( -31.90, z-score = -3.66, R) >dp4.chr3 9267907 118 - 19779522 ACACACACAGAAAUAAUGUUGGACUGUGCUUAAAGUAAAUAGUCUGGAUUGAUGAAAGUUA-UAUUCAAGAAUUCAUCAACAGGCAUUUACUUCAACUGCAGUUUAAACGAUAUGCAAC ............(((.(((((((((((..((.((((((((.(((((...(((((((..((.-......))..))))))).))))))))))))).))..))))))).)))).)))..... ( -29.20, z-score = -2.85, R) >droPer1.super_4 4590330 118 - 7162766 ACACACACAGAAAUAAUGUUGGACUGUGCUUAAAGUAAAUAGUCUGGAUUGAUGAAAGUUA-UAUUCAAGAAUUCAUCAACAGGCAUUUACUUCAACUGCAGUUUAAACGAUAUGCAAC ............(((.(((((((((((..((.((((((((.(((((...(((((((..((.-......))..))))))).))))))))))))).))..))))))).)))).)))..... ( -29.20, z-score = -2.85, R) >droWil1.scaffold_180697 3315652 113 + 4168966 -----AGAGAUCAUAAUGUUGAACUGUGCUUAAAGUAAAUAGUCUGGAUUGAUGAAAGUUA-UAUUCAAGAAUUCAUCAACAGGCAUUUACUUCAACUGCAGUUUGAACAAAAUGCAAC -----......(((..(((((((((((..((.((((((((.(((((...(((((((..((.-......))..))))))).))))))))))))).))..))))))).))))..))).... ( -30.00, z-score = -3.01, R) >droVir3.scaffold_12875 1241658 116 - 20611582 -GCGUCGCUCACAAA-UGUUGAACUGUGCUUAAAGUAAAUAGUCUGGAUUGAUGAAAGUUU-UAUUCAAGAAUUCAUCAACAGGCAUUUACUUCAACUGCAGUUUAAACGAAAUGCAAC -.....((.......-..(((((((((..((.((((((((.(((((...(((((((..(((-.....)))..))))))).))))))))))))).))..))))))))).......))... ( -31.89, z-score = -3.23, R) >droMoj3.scaffold_6496 4307040 118 - 26866924 -AUAUCGCGAAUAAACUGUUGAACUGUGCUUAAAGUAAAUAGUCUGGAUUGAUGAAAGUUUAUAUCUAAGAAUUCAUCAACAGGCAUUUACUUCAACUGCAGUUUAAACGAAAUGCAAC -.....(((.........(((((((((..((.((((((((.(((((...(((((((..((((....))))..))))))).))))))))))))).))..)))))))))......)))... ( -32.96, z-score = -4.31, R) >droGri2.scaffold_15245 8580541 111 + 18325388 -------AUAUCUAUCUGUUGAACUGUGCUUAAAGUAAAUAGUCUGGAUUGAUGAAAGUUU-UAUUCAAGAAUUCAUCAACAGGCAUUUACUUCAACUGCAGUUUAAACGAAAUGCAAC -------.......((..(((((((((..((.((((((((.(((((...(((((((..(((-.....)))..))))))).))))))))))))).))..)))))))))..))........ ( -30.60, z-score = -3.82, R) >consensus ____________ACAAUGUUGGACUGUGCUUAAAGUAAAUAGUCUGGAUUGAUGAAAGUUG_UAUUCAAGAAUUCAUCAACAGGCAUUUACUUCAACUGCAGUUUGAACGAAAUGCAAC ................(((((((((((..((.((((((((.(((((...(((((((..((........))..))))))).))))))))))))).))..))))))).))))......... (-29.95 = -29.43 + -0.52)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:10:01 2011