
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 5,152,699 – 5,152,757 |
| Length | 58 |
| Max. P | 0.591418 |

| Location | 5,152,699 – 5,152,757 |
|---|---|
| Length | 58 |
| Sequences | 8 |
| Columns | 60 |
| Reading direction | forward |
| Mean pairwise identity | 75.08 |
| Shannon entropy | 0.48413 |
| G+C content | 0.47943 |
| Mean single sequence MFE | -14.87 |
| Consensus MFE | -8.17 |
| Energy contribution | -8.57 |
| Covariance contribution | 0.41 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.533147 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
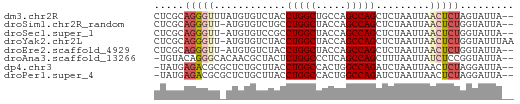
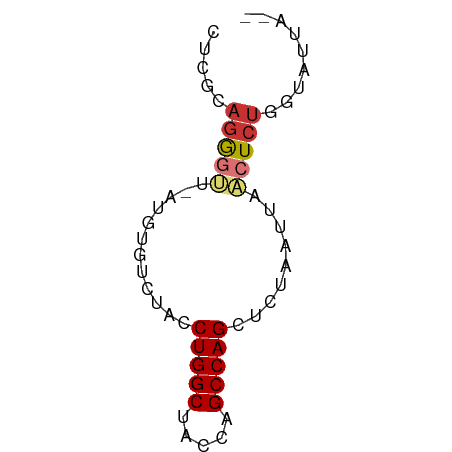
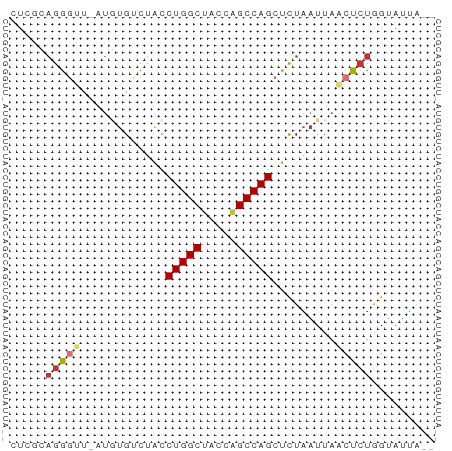
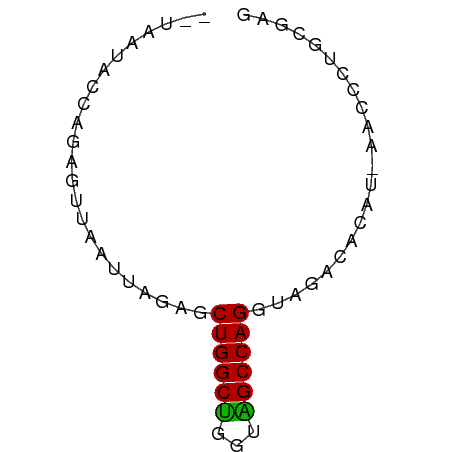
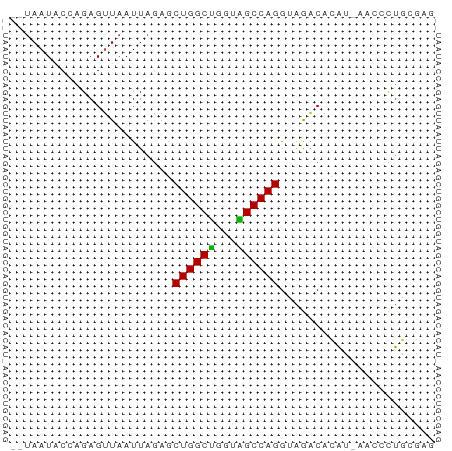
>dm3.chr2R 5152699 58 + 21146708 CUCGCAGGGUUUAUGUGUCUACCUGGCUGCCAGCCAGCUCUAAUUAACUCUAGUAUUA-- ...((((((((...((......(((((.....))))).....)).)))))).))....-- ( -13.10, z-score = -0.22, R) >droSim1.chr2R_random 1640330 57 + 2996586 CUCGCAGGGUU-AUGUGUCUGCCUGGCUACCAGCCAGCUCUAAUUAACUCUGGUAUUA-- ....(((((((-(.......((.((((.....))))))......))))))))......-- ( -17.72, z-score = -1.77, R) >droSec1.super_1 2784631 57 + 14215200 CUCGCAGGGUU-AUGUGUCCGCCUGGCUACCAGCCAGCUCUAAUUAACUCUGGUAUUA-- ....(((((((-(.......((.((((.....))))))......))))))))......-- ( -17.02, z-score = -1.50, R) >droYak2.chr2L 17807135 59 + 22324452 CUCGCAGGGUU-AUGUGUCUACCUGGCUACCAGCCAGCUCUAAUUAACUCUGGUAUUUAA ....(((((((-(.........(((((.....))))).......))))))))........ ( -16.69, z-score = -1.83, R) >droEre2.scaffold_4929 9135885 57 - 26641161 CUCGCAGGGUU-AUGUGUCUACCUGGCUACCAGCCAGCUCUAAUUAACUCUGGUAUUA-- ....(((((((-(.........(((((.....))))).......))))))))......-- ( -16.69, z-score = -1.81, R) >droAna3.scaffold_13266 11468467 57 - 19884421 -UGUACAGGGCACAACGCUACUCUGGCCCUCAGCCAGCUUUAAUUAUCUCCGGUAUUA-- -.((((.((((.....))....(((((.....)))))............)).))))..-- ( -11.70, z-score = -0.22, R) >dp4.chr3 9220849 57 + 19779522 -UAUGAGACGCGCUCUGCUUACCUGGCCACUGGCCAGAUCUAAUUAACUCUAGGAUUA-- -...(((..((.....))....((((((...))))))..........)))........-- ( -13.00, z-score = -1.03, R) >droPer1.super_4 4543972 57 + 7162766 -UAUGAGACGCGCUCUGCUUACCUGGCCACUGGCCAGAUCUAAUUAACUCUAGGAUUA-- -...(((..((.....))....((((((...))))))..........)))........-- ( -13.00, z-score = -1.03, R) >consensus CUCGCAGGGUU_AUGUGUCUACCUGGCUACCAGCCAGCUCUAAUUAACUCUGGUAUUA__ .....(((((............(((((.....))))).........)))))......... ( -8.17 = -8.57 + 0.41)
| Location | 5,152,699 – 5,152,757 |
|---|---|
| Length | 58 |
| Sequences | 8 |
| Columns | 60 |
| Reading direction | reverse |
| Mean pairwise identity | 75.08 |
| Shannon entropy | 0.48413 |
| G+C content | 0.47943 |
| Mean single sequence MFE | -13.90 |
| Consensus MFE | -9.10 |
| Energy contribution | -8.68 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.33 |
| Mean z-score | -0.87 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.591418 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
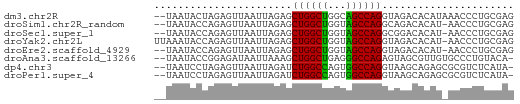
>dm3.chr2R 5152699 58 - 21146708 --UAAUACUAGAGUUAAUUAGAGCUGGCUGGCAGCCAGGUAGACACAUAAACCCUGCGAG --.....................(((((.....)))))((((...........))))... ( -10.70, z-score = 0.29, R) >droSim1.chr2R_random 1640330 57 - 2996586 --UAAUACCAGAGUUAAUUAGAGCUGGCUGGUAGCCAGGCAGACACAU-AACCCUGCGAG --.....((((.((((.......(((.((((...)))).))).....)-))).))).).. ( -14.20, z-score = -1.24, R) >droSec1.super_1 2784631 57 - 14215200 --UAAUACCAGAGUUAAUUAGAGCUGGCUGGUAGCCAGGCGGACACAU-AACCCUGCGAG --.....((((.((((.......(((.((((...)))).))).....)-))).))).).. ( -13.50, z-score = -0.55, R) >droYak2.chr2L 17807135 59 - 22324452 UUAAAUACCAGAGUUAAUUAGAGCUGGCUGGUAGCCAGGUAGACACAU-AACCCUGCGAG .......((((.((((.....(.(((((.....))))).).......)-))).))).).. ( -13.40, z-score = -1.01, R) >droEre2.scaffold_4929 9135885 57 + 26641161 --UAAUACCAGAGUUAAUUAGAGCUGGCUGGUAGCCAGGUAGACACAU-AACCCUGCGAG --.....((((.((((.....(.(((((.....))))).).......)-))).))).).. ( -13.40, z-score = -1.03, R) >droAna3.scaffold_13266 11468467 57 + 19884421 --UAAUACCGGAGAUAAUUAAAGCUGGCUGAGGGCCAGAGUAGCGUUGUGCCCUGUACA- --...(((.((..(((((.....(((((.....)))))......)))))..)).)))..- ( -14.60, z-score = -0.48, R) >dp4.chr3 9220849 57 - 19779522 --UAAUCCUAGAGUUAAUUAGAUCUGGCCAGUGGCCAGGUAAGCAGAGCGCGUCUCAUA- --........(((........((((((((...))))))))..((.....))..)))...- ( -15.70, z-score = -1.47, R) >droPer1.super_4 4543972 57 - 7162766 --UAAUCCUAGAGUUAAUUAGAUCUGGCCAGUGGCCAGGUAAGCAGAGCGCGUCUCAUA- --........(((........((((((((...))))))))..((.....))..)))...- ( -15.70, z-score = -1.47, R) >consensus __UAAUACCAGAGUUAAUUAGAGCUGGCUGGUAGCCAGGUAGACACAU_AACCCUGCGAG .......................((((((...))))))...................... ( -9.10 = -8.68 + -0.42)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:09:53 2011