| Sequence ID | dm3.chr2R |
|---|---|
| Location | 5,087,535 – 5,087,643 |
| Length | 108 |
| Max. P | 0.907536 |

| Location | 5,087,535 – 5,087,643 |
|---|---|
| Length | 108 |
| Sequences | 11 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 66.91 |
| Shannon entropy | 0.68727 |
| G+C content | 0.49145 |
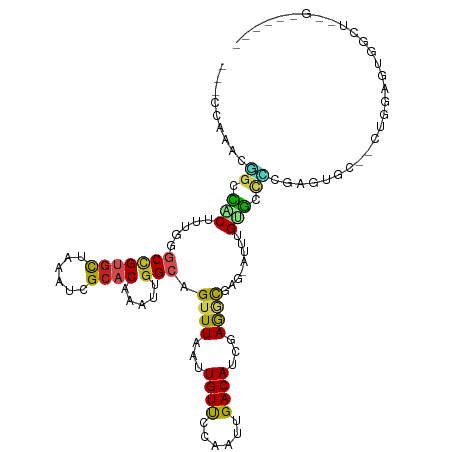
| Mean single sequence MFE | -33.75 |
| Consensus MFE | -10.41 |
| Energy contribution | -9.83 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.70 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.907536 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 5087535 108 - 21146708 -CUCCAAACGGCCACUUUGGGCCGUGCUAAAUCGCACAAAUUGGCAGUUUAAUUGUUCCAAUUGACAUCGAGGCGAG-AUUUGUGCCCCGGUUGC--CUGGAGUGGCCUCGA----- -........((((((((..(((.((((......)))).((((((((((...))))..))))))...((((.((((..-.....)))).)))).))--)..))))))))....----- ( -42.20, z-score = -2.83, R) >droGri2.scaffold_15245 12116711 108 + 18325388 ---UCAGUUUGAGUCU-----CUGUAUCGAAUCGCCCAAAUUGGCAGUUUAAUUGUUCCAAUUGACACCAAAUCUUGUUUCGGACCCUCGAGUGUUGCCCAAAUAAAUGGGCAAUG- ---....((((((..(-----(((...(((...(((......))).(((.(((((...))))))))........)))...))))..)))))).((((((((......)))))))).- ( -29.10, z-score = -2.49, R) >droMoj3.scaffold_6496 264845 104 - 26866924 -------UCGGGGCCU----GCCGUGUUAAAUCGCACAAAUUGGCAGUUUAAUUGUUCCAAUUGACACCAAAUUUUGUUUU-GGCCCUUAAGUGGC-CCAGAAUAAAUGGGCAACUG -------..(((((((----(((((((......)))).....))))(((.(((((...))))))))...............-))))))..(((.((-(((.......))))).))). ( -31.50, z-score = -1.38, R) >droVir3.scaffold_12823 450474 101 - 2474545 -------CCAGGGCCU-----CCGUGCCGAAUUGCCCAAAUUGGUAGUUUAAUUGUUCCAAUUGACACCGAAACUUGUUU--GUCCCUCGAGUGGC-CCAAAAUAAAUGGGCAAUG- -------....(((..-----....)))..((((((((..(((((.(((.(((((...)))))))))))))......(((--(..((......)).-.)))).....)))))))).- ( -27.90, z-score = -1.50, R) >droPer1.super_4 2035531 93 + 7162766 CUGUUAGGUCGGUCCUCUGGGCCGUGUUAAAUCGCACAAAUUGGCAGUUUAAUUGUUCCAAUUGACAUCGAAACGAGUCCCCGAGCAGUGCCU------------------------ (((((((...(((((...)))))((((......))))...)))))))....(((((((.....(((.(((...))))))...)))))))....------------------------ ( -24.60, z-score = -0.77, R) >dp4.chr3 9968367 93 + 19779522 CUGUUAGGUCGGUCCUCUGGGCCGUGUUAAAUCGCACAAAUUGGCAGUUUAAUUGUUCCAAUUGACAUCGAAACGAGUCCCCGAGCACUGCCU------------------------ .....(((.((((.(((.(((..((((......)))).....(((.((((...((((......))))...))))..))))))))).)))))))------------------------ ( -25.00, z-score = -0.93, R) >droAna3.scaffold_13266 11397511 102 + 19884421 ---CCAGACGGCCACUUUCGGCCGUGCUAAAUCGCACAAAUUGGCAGUUUAAUUGUCCCAAUUGACAUUGAGGCAAG-AUUUGUGCUCUGGGUCC--CUUAAGUACCU--------- ---((((((((((......))))))........(((((((((....((((...((((......))))...))))..)-)))))))).))))....--...........--------- ( -33.70, z-score = -2.66, R) >droEre2.scaffold_4929 9071815 105 + 26641161 ---CCAAACGGCCACUUUGGGCCGUGUUAAAUCGCACAAAUUGGCAGUUUAAUUGUUCCAAUUGACAUCGAGGCGAG-AUUUGUGCCCGGAGUCC--UUGGAGUGGCCCUG------ ---......((((((((..((..((((......)))).((((((((((...))))..))))))(((..((.((((..-.....))))))..))))--)..))))))))...------ ( -37.20, z-score = -2.13, R) >droYak2.chr2L 17742341 105 - 22324452 ---CCAAAUGGCCACUUUGGGCCGUGCUAAAUCGCACAAAUUAGCAGUUUAAUUGUUCCAAUUGACAUCGAGGCGAG-AUUUGUGCCCGGAGUCC--CUGGAGUGGCUCUA------ ---......((((((((..((..((((......)))).....((((((...))))))......(((..((.((((..-.....))))))..))))--)..))))))))...------ ( -35.60, z-score = -2.01, R) >droSec1.super_1 2719849 108 - 14215200 -CUCCAAACGGCCACUUUGGGCCGUGCUAAAUCGCACAAAUUGGCAGUUUAAUUGUUCCAAUUGACAUCGAGGCGAG-AUUUGUGCCCCGGUUGC--CUGGAGUGGCCUCGA----- -........((((((((..(((.((((......)))).((((((((((...))))..))))))...((((.((((..-.....)))).)))).))--)..))))))))....----- ( -42.20, z-score = -2.83, R) >droSim1.chr2R 3750793 108 - 19596830 -CUCCAAACGGCCACUUUGGGCCGUGCUAAAUCGCACAAAUUGGCAGUUUAAUUGUUCCAAUUGACAUCGAGGCGAG-AUUUGUGCCCCGGUUGC--CUGGAGUGGCCUCGA----- -........((((((((..(((.((((......)))).((((((((((...))))..))))))...((((.((((..-.....)))).)))).))--)..))))))))....----- ( -42.20, z-score = -2.83, R) >consensus ___CCAAACGGCCACUUUGGGCCGUGCUAAAUCGCACAAAUUGGCAGUUUAAUUGUUCCAAUUGACAUCGAGGCGAG_AUUUGUGCCCCGAGUGC__CUGGAGUGGCU__G______ .........((.(((.....(((((((......)))).....))).((((...((((......))))...))))........))).))............................. (-10.41 = -9.83 + -0.58)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:09:45 2011