| Sequence ID | dm3.chr2R |
|---|---|
| Location | 5,029,042 – 5,029,157 |
| Length | 115 |
| Max. P | 0.513330 |

| Location | 5,029,042 – 5,029,157 |
|---|---|
| Length | 115 |
| Sequences | 9 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 73.28 |
| Shannon entropy | 0.55600 |
| G+C content | 0.42803 |
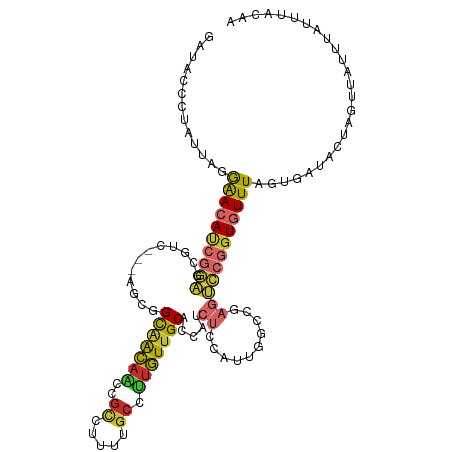
| Mean single sequence MFE | -25.60 |
| Consensus MFE | -13.60 |
| Energy contribution | -13.59 |
| Covariance contribution | -0.01 |
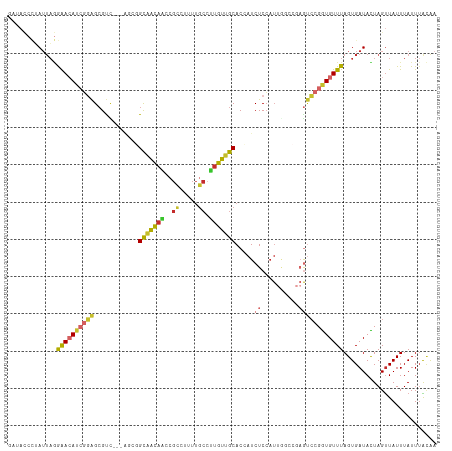
| Combinations/Pair | 1.57 |
| Mean z-score | -0.87 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.513330 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 5029042 115 + 21146708 GAUACCCUAUUAGGAACAUCGGAGCGUUGUCACUGGCAACAACCGCCUUUUGCCUUGUUGCACCAUCUCCAUUGGACGAGUCCGGUGUUUAGUGAUACUAGUUAUUUAUUUACGA .......(((((.((((((((((.(((........(((((((..((.....)).))))))).(((.......))))))..))))))))))..))))).................. ( -29.20, z-score = -1.08, R) >droSim1.chr2R 3697991 115 + 19596830 GAUACCCUAUUAGGAACAUCGGAGCGUCGUCAGCGGCAACAACCGCCUUUUGCCUUGUUGCACCAUCUCCAUUGGCCGAGUCCGGUGUUUAGUGAUACUAGUUAUUUAUUUACAA .......(((((.((((((((((.(.(((......(((((((..((.....)).))))))).(((.......))).))))))))))))))..))))).................. ( -27.90, z-score = -0.59, R) >droSec1.super_1 2668128 114 + 14215200 UGUACCCUAUUAGGAACAUCGGAGCGUCGUCAUCGGCAACAACCGUCUUUUGC-UUGUUGCACCAUCUCCAUUGGCCGAGUCCGGUGUUUAGUGAUACUAGUUAUUUAUUUACAA .......(((((.((((((((((.(.(((......(((((((.((.....)).-))))))).(((.......))).))))))))))))))..))))).................. ( -28.40, z-score = -1.04, R) >droYak2.chr2L 17690816 112 + 22324452 GAUACCCUAUUAGGAACAUCGGAGCGUC---AGCGGCAACAACCGCCUUUUGCCUUGUUGCACCAUCUCCAUUGGCCGAGUCCGGUGUUUAGUGAUACUAGUUAUUUAUUUACAA .......(((((.((((((((((((...---.)).(((((((..((.....)).))))))).(((.......))).....))))))))))..))))).................. ( -27.50, z-score = -0.82, R) >droEre2.scaffold_4929 9020419 112 - 26641161 GAUACCCUAUUAGGAACAUCGGAGCGUC---AGCGGCAACAACCGCCUUUUGCCUUGUUGCACCAUCUCCAUUGGCCGAGUCCGGUGUUUAGUGAUACUAGUUAUUUAUUUACAA .......(((((.((((((((((((...---.)).(((((((..((.....)).))))))).(((.......))).....))))))))))..))))).................. ( -27.50, z-score = -0.82, R) >droAna3.scaffold_13266 13145613 112 - 19884421 GUAAUCCUAUUAGGAACAUCGGAGCGCU---AGAUGCAACAGUAGCCUUUUGCCUUGUUGCACAAUCUCCAUUAGCCGAGUCCGGUGUUUAGUGAUAUUAGUUAUUUAUUUAAGU (((((..(((((.((((((((((.((((---(((((((((((..((.....)).))))))))...))).....)).))..))))))))))..)))))...))))).......... ( -27.30, z-score = -1.51, R) >droWil1.scaffold_181141 1429219 101 - 5303230 AAAACCUGAUUAGAAACAUCGGAGAGUGCUCGC--GCUGUGACUAUAU--------AUUGCACAAUCUCCCUAG----AAUCCGGUGUUUAGUGAUAUUAGUUAUUUAUUGACAA ........((((.((((((((((.((((....)--)))(((.(.....--------...))))...........----..))))))))))..))))....((((.....)))).. ( -22.10, z-score = -1.18, R) >droVir3.scaffold_12875 13332247 94 + 20611582 -------AAUUAAAGAGACAUCGGCGCU-------GUGACAGACGUGUUUUGCCCUGUUGCACAAUCUCGAA-------GCGCGGUGUUUAGUGAUAUUAGUUAUUUAUUUGUAA -------.((((...((((((((.((((-------(..((((..((.....)).))))..).(......).)-------)))))))))))..))))................... ( -22.40, z-score = -0.40, R) >droMoj3.scaffold_6496 20628683 94 + 26866924 -------AAAAUUAGAAACAUCGGCGCU-------GUGACAGACGUGUUUUGCCCUGUUGCACAAUCUCCAU-------AACCGAUGUUUAGUGAUAUUAGCUAUUUAUUUGUAA -------...((((.(((((((((...(-------(..((((..((.....)).))))..))..........-------..)))))))))..))))................... ( -18.06, z-score = -0.42, R) >consensus GAUACCCUAUUAGGAACAUCGGAGCGUC___AGCGGCAACAACCGCCUUUUGCCUUGUUGCACCAUCUCCAUUGGCCGAGUCCGGUGUUUAGUGAUACUAGUUAUUUAUUUACAA .............((((((((((............(((((((..((.....)).)))))))..........((....)).))))))))))......................... (-13.60 = -13.59 + -0.01)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:09:43 2011