| Sequence ID | dm3.chr2R |
|---|---|
| Location | 5,028,580 – 5,028,683 |
| Length | 103 |
| Max. P | 0.774611 |

| Location | 5,028,580 – 5,028,682 |
|---|---|
| Length | 102 |
| Sequences | 7 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 65.99 |
| Shannon entropy | 0.62443 |
| G+C content | 0.52872 |
| Mean single sequence MFE | -23.47 |
| Consensus MFE | -9.46 |
| Energy contribution | -10.21 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.730407 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
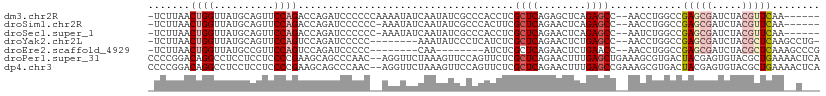
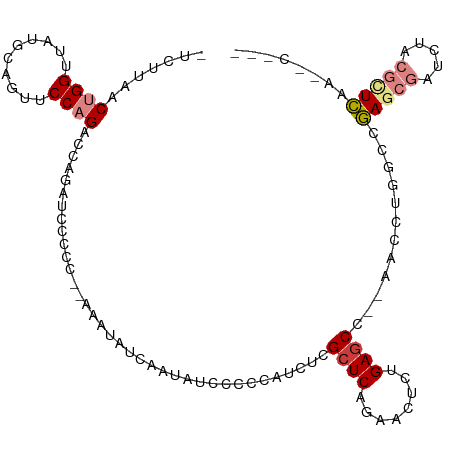
>dm3.chr2R 5028580 102 - 21146708 -CUUAACUGGUUAUGCAGUUCCAGACCAGAUCCCCCCAAAAUAUCAAUAUCGCCCACCUCGCUCAGAGCUCAGAGCC--AACCUGGCCGAGCGAUCUACGUUCAA------- -.....((((((.((......))))))))......................(((......((((........)))).--.....))).(((((.....)))))..------- ( -21.10, z-score = -1.29, R) >droSim1.chr2R 3697527 101 - 19596830 -CUUAACUGGUUAUGCAGUUCCAGACCAGAUCCCCCC-AAAUAUCAAUAUCGCCCACUUCGCUCAGAACUCAGAGCC--AACCUGGCCGAGCGAUCUACGUUCAA------- -.....((((((.((......))))))))........-.............(((......((((........)))).--.....))).(((((.....)))))..------- ( -21.10, z-score = -1.68, R) >droSec1.super_1 2667663 101 - 14215200 -CUUAACUGGUUAUGCAGUUCCAGACCAGAUCCCCCC-AAAUAUCAAUAUCGCCCACCUCGCUCAGAACUCAGAGCC--AAUCUGGCCGAGCGAUCUACGUUCAA------- -.....((((((.((......))))))))........-.............(((......((((........)))).--.....))).(((((.....)))))..------- ( -21.10, z-score = -1.61, R) >droYak2.chr2L 17690364 99 - 22324452 -CUUAACUGGUUAUGCAGUUCCAGUCCAGAUCCCCC--------AAAUAUCCCUCAUCUCGCUCAGAACUCUGAGCC--AACCUGGCCGAGCGAUCUACGCUCAAGCCUG-- -((..(((((..........)))))..)).......--------................((((((....)))))).--.....(((.(((((.....)))))..)))..-- ( -26.70, z-score = -3.09, R) >droEre2.scaffold_4929 9019973 92 + 26641161 -CUUAACUGGUUAUGCCGUUCCAGUCCAGAUCCCCC--------CAA--------AUCUCGCUCAGAACUCUGAACC--AACCUGGCCGAGCGAUCUACGCUCAAAGCCCG- -......(((((.....((((.(((..((((.....--------...--------)))).)))..))))....))))--)....(((.(((((.....)))))...)))..- ( -20.30, z-score = -1.89, R) >droPer1.super_31 628426 110 + 935084 CCCGGACAGGCCUCCUCCUCCCCGAAGCAGCCCAAC--AGGUUCUAAAGUUCCAGUUCUCGCUCAGAACUUUGAGCUGAAAGCGUGACUACGAGUGUACGCUGAAAACUCAU ..(((..(((......)))..)))...((((.(((.--.((..(....)..))((((((.....))))))))).))))..((((((((.....)).)))))).......... ( -26.30, z-score = 0.10, R) >dp4.chr3 17484919 110 - 19779522 CCCGGACAGGCCUCCUCCUCCCCGAAGCAGCCCAAC--AGGUUCUAAAGUUCCAGUUCUCGCUCAGAACUUUGAGCCGAAAGCGUGACUACGAGUGUACGCUGAAAACUCAU ..(((..(((......)))..)))............--.(((((.(((((((.(((....)))..))))))))))))...((((((((.....)).)))))).......... ( -27.70, z-score = -0.48, R) >consensus _CUUAACUGGUUAUGCAGUUCCAGACCAGAUCCCCC__AAAUAUCAAUAUCCCCCAUCUCGCUCAGAACUCUGAGCC__AACCUGGCCGAGCGAUCUACGCUCAA__C____ ......((((..........))))....................................((((........))))............(((((.....)))))......... ( -9.46 = -10.21 + 0.76)
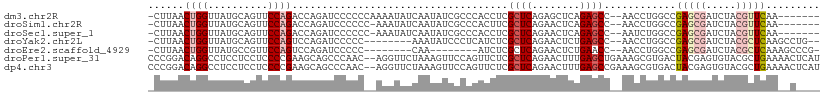
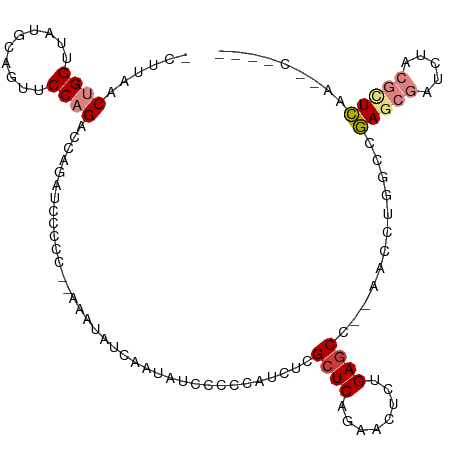
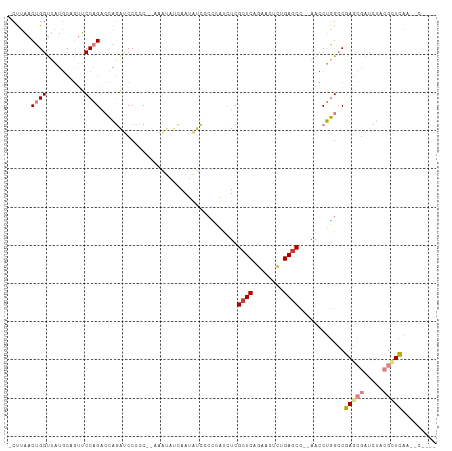
| Location | 5,028,580 – 5,028,683 |
|---|---|
| Length | 103 |
| Sequences | 7 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 66.14 |
| Shannon entropy | 0.62443 |
| G+C content | 0.52756 |
| Mean single sequence MFE | -23.79 |
| Consensus MFE | -9.46 |
| Energy contribution | -10.21 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.774611 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 5028580 103 - 21146708 -UCUUAACUGGUUAUGCAGUUCCAGACCAGAUCCCCCCAAAAUAUCAAUAUCGCCCACCUCGCUCAGAGCUCAGAGCC--AACCUGGCCGAGCGAUCUACGUUCAA------ -......((((((.((......))))))))......................(((......((((........)))).--.....))).(((((.....)))))..------ ( -21.10, z-score = -1.26, R) >droSim1.chr2R 3697527 102 - 19596830 -UCUUAACUGGUUAUGCAGUUCCAGACCAGAUCCCCCC-AAAUAUCAAUAUCGCCCACUUCGCUCAGAACUCAGAGCC--AACCUGGCCGAGCGAUCUACGUUCAA------ -......((((((.((......))))))))........-.............(((......((((........)))).--.....))).(((((.....)))))..------ ( -21.10, z-score = -1.65, R) >droSec1.super_1 2667663 102 - 14215200 -UCUUAACUGGUUAUGCAGUUCCAGACCAGAUCCCCCC-AAAUAUCAAUAUCGCCCACCUCGCUCAGAACUCAGAGCC--AAUCUGGCCGAGCGAUCUACGUUCAA------ -......((((((.((......))))))))........-.............(((......((((........)))).--.....))).(((((.....)))))..------ ( -21.10, z-score = -1.61, R) >droYak2.chr2L 17690364 100 - 22324452 -UCUUAACUGGUUAUGCAGUUCCAGUCCAGAUCCCCC--------AAAUAUCCCUCAUCUCGCUCAGAACUCUGAGCC--AACCUGGCCGAGCGAUCUACGCUCAAGCCUG- -(((..(((((..........)))))..)))......--------................((((((....)))))).--.....(((.(((((.....)))))..)))..- ( -28.10, z-score = -3.52, R) >droEre2.scaffold_4929 9019973 93 + 26641161 -UCUUAACUGGUUAUGCCGUUCCAGUCCAGAUCCCCC--------CAA--------AUCUCGCUCAGAACUCUGAACC--AACCUGGCCGAGCGAUCUACGCUCAAAGCCCG -(((..(((((..........)))))..)))......--------...--------.......((((....))))...--.....(((.(((((.....)))))...))).. ( -21.10, z-score = -2.18, R) >droPer1.super_31 628427 110 + 935084 CCCCGGACAGGCCUCCUCCUCCCCGAAGCAGCCCAAC--AGGUUCUAAAGUUCCAGUUCUCGCUCAGAACUUUGAGCUGAAAGCGUGACUACGAGUGUACGCUGAAAACUCA ...(((..(((......)))..)))...((((.(((.--.((..(....)..))((((((.....))))))))).))))..((((((((.....)).))))))......... ( -26.30, z-score = 0.22, R) >dp4.chr3 17484920 110 - 19779522 CCCCGGACAGGCCUCCUCCUCCCCGAAGCAGCCCAAC--AGGUUCUAAAGUUCCAGUUCUCGCUCAGAACUUUGAGCCGAAAGCGUGACUACGAGUGUACGCUGAAAACUCA ...(((..(((......)))..)))............--.(((((.(((((((.(((....)))..))))))))))))...((((((((.....)).))))))......... ( -27.70, z-score = -0.36, R) >consensus _UCUUAACUGGUUAUGCAGUUCCAGACCAGAUCCCCC__AAAUAUCAAUAUCCCCCAUCUCGCUCAGAACUCUGAGCC__AACCUGGCCGAGCGAUCUACGCUCAA__C___ .......((((..........))))....................................((((........))))............(((((.....)))))........ ( -9.46 = -10.21 + 0.76)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:09:42 2011