
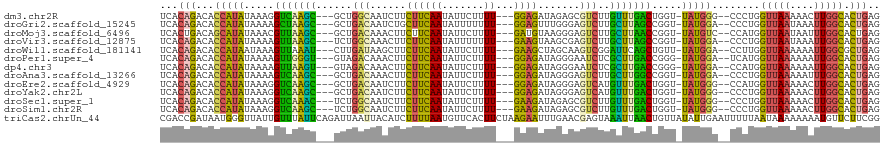
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 4,984,560 – 4,984,671 |
| Length | 111 |
| Max. P | 0.982434 |

| Location | 4,984,560 – 4,984,671 |
|---|---|
| Length | 111 |
| Sequences | 13 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.85 |
| Shannon entropy | 0.45535 |
| G+C content | 0.39207 |
| Mean single sequence MFE | -31.72 |
| Consensus MFE | -15.43 |
| Energy contribution | -13.65 |
| Covariance contribution | -1.78 |
| Combinations/Pair | 1.78 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.10 |
| SVM RNA-class probability | 0.982434 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 4984560 111 + 21146708 UCACAGACACCAUAUAAAGGUCAAGC---GCUGGCAAUCUUCUUCAAUAUUCUUUU---GGAGAUAGAGCGUCUUGUUUGACUGGU-UAUGGG--CCCUGGUUAAAAACUUGGCACUGAG ...(((...(((((....((((((((---(..(((..((((((((((.......))---))))).)))..))).)))))))))...-)))))(--((..(((.....))).))).))).. ( -37.50, z-score = -3.43, R) >droGri2.scaffold_15245 10600999 111 + 18325388 UCACAGACACCAUAUAAAAGCUAAGC---GCUGACAAUCUGCUUCAAUAUUUUUUU---GGAGUUUGGGAGUCUUGCUUAGCCGGU-UAUGGA--CCCUGGUUAAUAAAUUGGCACUGAG ...(((...(((((.....(((((((---(..(((..((.(((((((.......))---)))))..))..))).))))))))....-))))).--..(..(((....)))..)..))).. ( -32.10, z-score = -2.16, R) >droMoj3.scaffold_6496 5124798 111 + 26866924 UCACUGACAGCAUAUAAACGUUAAGC---GCUGACAAACUUCUUCAAUAUUCUUUU---GAUGUAAGGGAGUCUUGCUUAACCGGU-UAUGUC--CCAUGGUUAAUAAUUUGGCACUGAG .......(((.((((((..(((((((---(..(((...((((.((((.......))---)).).)))...))).))))))))...)-))))).--.....(((((....))))).))).. ( -22.80, z-score = -0.38, R) >droVir3.scaffold_12875 20244594 111 - 20611582 UCACAGACACCAUAUAAAAGUUAAGC---UCUGGCAAACUUCUUCAAUAUUUUUUU---GAAGUAAGCGAGUCUUGCUUAGCCGGU-UAUGGA--CCCUGGUUAAUAAAUUGGCACUGAG ...(((...............(((((---...(((...(((((((((.......))---)))).)))...)))..)))))((((((-(...((--(....)))....))))))).))).. ( -27.90, z-score = -1.76, R) >droWil1.scaffold_181141 3774418 111 + 5303230 UCACAGACACCAUAAUAAAGUUAAAU---CUUGAUAAGCUUCUUCAAUAUUCUUUU---GAAGCUAGCAAGUCGGAUUCAGCUGUU-UAUGGA--CCUUGGUUAAAAAAUUGGCGCUGAG ...(((.(.((((((...((((.(((---((.(((..(((.((((((.......))---))))..)))..)))))))).))))..)-))))).--.....(((((....))))))))).. ( -28.90, z-score = -2.54, R) >droPer1.super_4 5021358 111 + 7162766 UCACAGACACCAUAUAAAAGUUGGGU---GUAGACAAACUUCUUCAAUAUUCUUUU---GGAGAUAGGGAAUCUCGCUUGACCGGG-UAUGGA--UCAUGGUUAAAAAAUUGGCACUGAG ...(((...((((((....((..(((---(.(((....(((((((((.......))---))))).))....)))))))..))...)-))))).--.....(((((....))))).))).. ( -29.60, z-score = -2.08, R) >dp4.chr3 9696655 111 + 19779522 UCACAGACACCAUAUAAAAGUUAAGU---GUAGACAAACUUCUUCAAUAUUCUUUU---GGAGAUAGGGAAUCUCGCUUGACCGGG-UAUGGA--CCAUGGUUAAAAAAUUGGCACUGAG ...(((...((((((....(((((((---(.(((....(((((((((.......))---))))).))....)))))))))))...)-))))).--.....(((((....))))).))).. ( -29.90, z-score = -2.48, R) >droAna3.scaffold_13266 16370958 111 - 19884421 UCACAGACACCAUAUAAAAGUCAAGC---GCUGACAAACUUCUUCAAUAUUCUUUU---GGAGAUAGGGAGUCUUGCUUGGCCGGU-UAUGGA--CCCUGGUUAAAAAUUUGGCACUGAG ...(((...(((((.....(((((((---(..(((...(((((((((.......))---))))).))...))).))))))))....-))))).--.....(((((....))))).))).. ( -30.80, z-score = -1.81, R) >droEre2.scaffold_4929 8976156 111 - 26641161 UCACAGACACCAUAUAAAGGUCAAGC---GCUGACAAUCUUCUUCAAUAUUCUUUU---GGAGAUAGGGAGUCAUGUUUGACUGGU-UAUGGG--CCAUGGUUAAAAACUUGGCACUGAG ...(((...(((((....((((((((---(.((((..((((((((((.......))---))))).)))..)))))))))))))...-)))))(--(((.(((.....))))))).))).. ( -40.30, z-score = -4.84, R) >droYak2.chr2L 17646651 111 + 22324452 UCACAGACACCAUAUAAAGGUCAAGC---GCUGACAAUCUUCUUCAAUAUUCUUUU---GGAGAUAGGGAGUCAUGUUUGACUGGU-UAUGGG--CCCUGGUUAAAAACUUGGCACUGAG ...(((...(((((....((((((((---(.((((..((((((((((.......))---))))).)))..)))))))))))))...-)))))(--((..(((.....))).))).))).. ( -39.30, z-score = -4.12, R) >droSec1.super_1 2622783 111 + 14215200 UCACAGACACCAUAUAAAGGUCAAAC---UCUGGCAAUCUUCUUCAAUAUUCUUUU---GAAGAUAGAGCGUCUUGUUUGACUGGU-UAUGGG--CCCUGGUUAAAAACUUGGCACUGAG ...(((...(((((....((((((((---...(((..((((((((((.......))---))))).)))..)))..))))))))...-)))))(--((..(((.....))).))).))).. ( -37.80, z-score = -4.39, R) >droSim1.chr2R 3655171 111 + 19596830 UCACAGACACCAUAUAAAGGUCAAGC---UCUGGCAAUCUUCUUCAAUAUUCUUUU---GAAGAUAGAGCGUCUUGUUUGACUGGU-UAUGGG--CCCUGGUUAAAAACUUGGCACUGAG ...(((...(((((....((((((((---...(((..((((((((((.......))---))))).)))..)))..))))))))...-)))))(--((..(((.....))).))).))).. ( -37.80, z-score = -3.94, R) >triCas2.chrUn_44 114464 120 + 525920 CGACCGAUAAUGGGUUAUUGUUUAUUCAGAUUAAUUACAUCUUUUAAUGUUCACUUCUAAGAAUUUGAACGAGUAAAUUAACUGUUAUAUUGAAUUUUUAAUAAAAAAAAUGUUCUUCGG ...((((.....(((((..((((((((((((.......))))......(((((.(((...)))..))))))))))))))))))....((((((....)))))).............)))) ( -17.70, z-score = -0.12, R) >consensus UCACAGACACCAUAUAAAAGUUAAGC___GCUGACAAUCUUCUUCAAUAUUCUUUU___GGAGAUAGGGAGUCUUGCUUGACUGGU_UAUGGA__CCCUGGUUAAAAAAUUGGCACUGAG ...(((...(((((.....(((((((......(((......((((..............)))).......)))..))))))).....)))))........(((((....))))).))).. (-15.43 = -13.65 + -1.78)
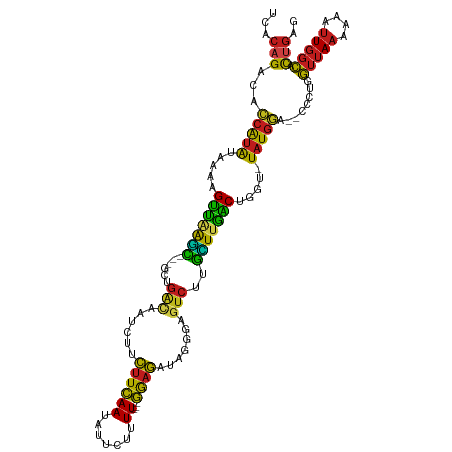
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:09:38 2011