
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 4,961,320 – 4,961,378 |
| Length | 58 |
| Max. P | 0.980269 |

| Location | 4,961,320 – 4,961,378 |
|---|---|
| Length | 58 |
| Sequences | 11 |
| Columns | 68 |
| Reading direction | reverse |
| Mean pairwise identity | 81.24 |
| Shannon entropy | 0.34024 |
| G+C content | 0.33602 |
| Mean single sequence MFE | -13.80 |
| Consensus MFE | -9.17 |
| Energy contribution | -9.06 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.980269 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
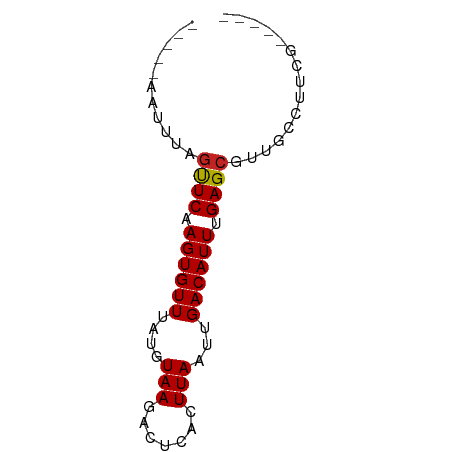
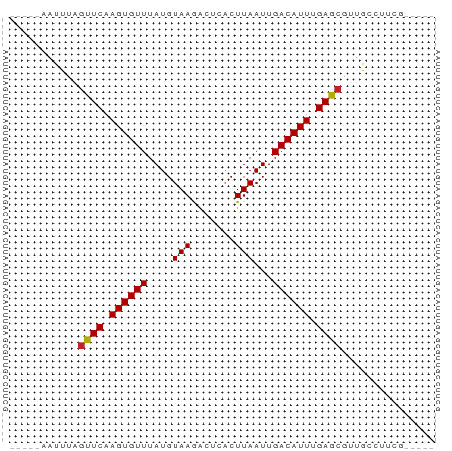
>dm3.chr2R 4961320 58 - 21146708 -----AAUUUAGUUCAAGUGUUUAUGUAAGACUCACUUAAUUGACAUUAGAGCGUUACAUUCG----- -----......((((.((((((....((((.....))))...)))))).))))..........----- ( -11.40, z-score = -1.89, R) >droSim1.chr2R 3631575 57 - 19596830 ------AUUUAGUUCAAGUGUUUAUGUAAGACUCACUUAAUUGACAUUAGAGCGUUACAUUCG----- ------.....((((.((((((....((((.....))))...)))))).))))..........----- ( -11.40, z-score = -1.95, R) >droSec1.super_1 2599608 58 - 14215200 -----AAUUUAGUUCAAGUGUUUAUGUAAGACUCACUUAAUUGACAUUAGAGCGUUACAUUCG----- -----......((((.((((((....((((.....))))...)))))).))))..........----- ( -11.40, z-score = -1.89, R) >droYak2.chr2L 17622452 58 - 22324452 -----AAUUUAGUUCAAGUGUUUAUGUAAGACUCACUUAAUUGACAUUAGAGCGUUACAUUCA----- -----......((((.((((((....((((.....))))...)))))).))))..........----- ( -11.40, z-score = -1.96, R) >droEre2.scaffold_4929 8952765 58 + 26641161 -----AAUUUAGUUCAAGUGUUUAUGUAAGAUUCACUUAAUUGACAUUAGAGCGUUACAUUCA----- -----......((((.((((((....((((.....))))...)))))).))))..........----- ( -11.40, z-score = -2.34, R) >droAna3.scaffold_13266 16349383 63 + 19884421 -----AAUUUGGUUCAAGUGUUUAUGUAAGGCCCACUUAAUUGACAUUUGAGAGCGGUCAUAGUUACA -----...(((.((((((((((....((((.....))))...))))))))))..)))........... ( -13.00, z-score = -1.03, R) >dp4.chr3 9674731 63 - 19779522 AAUUUGAUUUAGUUCAAGUGUUUAUGUAAGACUCUCUUAAUUGACAUUUGAGCGCUGCCGUCG----- .....(((...(((((((((((....((((.....))))...)))))))))))......))).----- ( -16.60, z-score = -2.95, R) >droPer1.super_4 4999446 63 - 7162766 AAUUUAAUUUAGUUCAAGUGUUUAUGUAAGACUCUCUUAAUUGACAUUUGAGCGCUGCCUUCG----- ...........(((((((((((....((((.....))))...)))))))))))..........----- ( -16.50, z-score = -3.77, R) >droMoj3.scaffold_6496 5540553 55 - 26866924 -----AAUUUGGCUCAAGUGUUUAUGUAAAGUCCACUUAAUUGACAUUUGAGCGUUGUCG-------- -----......(((((((((((...((.(((....))).)).))))))))))).......-------- ( -16.50, z-score = -3.78, R) >droGri2.scaffold_15245 10971164 55 - 18325388 -----AAUUUGGCUCAAGUGUUUAUGUAAAGCCCACUUAAUUGACAUUUGAGCGCUGUCG-------- -----......(((((((((((...((.(((....))).)).))))))))))).......-------- ( -15.70, z-score = -2.87, R) >droVir3.scaffold_12875 31370 55 + 20611582 -----AAUUUGGCUCAAGUGUUUAUGUAAAGUCCACUUAAUUGACAUUUGAGCGUUGUCG-------- -----......(((((((((((...((.(((....))).)).))))))))))).......-------- ( -16.50, z-score = -3.78, R) >consensus _____AAUUUAGUUCAAGUGUUUAUGUAAGACUCACUUAAUUGACAUUUGAGCGUUGCCUUCG_____ ...........((((.((((((....(((.......)))...)))))).))))............... ( -9.17 = -9.06 + -0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:09:34 2011