
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 4,919,946 – 4,920,024 |
| Length | 78 |
| Max. P | 0.839142 |

| Location | 4,919,946 – 4,920,024 |
|---|---|
| Length | 78 |
| Sequences | 11 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 70.65 |
| Shannon entropy | 0.55782 |
| G+C content | 0.47581 |
| Mean single sequence MFE | -20.70 |
| Consensus MFE | -11.52 |
| Energy contribution | -11.43 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.13 |
| Mean z-score | -0.74 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.514303 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
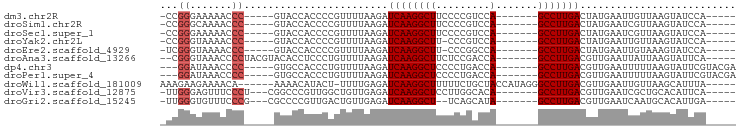
>dm3.chr2R 4919946 78 + 21146708 -----UGGAUACUUAACAAUUCAUAGUCAAGGC-------UGGACGGGGAAGCCUUGAUCUUAAAACGGGGUGGUAC-----GGGUUUUUCCCGG- -----....((((...(.......(((((((((-------(.........)))))))).)).......)...))))(-----(((.....)))).- ( -21.74, z-score = -0.84, R) >droSim1.chr2R 3589706 78 + 19596830 -----UGGAUACUUAACGAUUCAUAGUCAAGGC-------UGGACGGGGAAGCCUUGAUCUUAAAACGGGGUGGUAC-----GGGUUUUGCCCGG- -----....((((...((.((.(..((((((((-------(.........)))))))))..).)).))....))))(-----(((.....)))).- ( -23.70, z-score = -1.21, R) >droSec1.super_1 2558733 78 + 14215200 -----UGGAUACUUAACGAUUCAUAGUCAAGGC-------UGGACGGGGAAGCCUUGAUCUUAAAACGGGGUGGUAC-----GGGUUUUUCCCGG- -----....((((...((.((.(..((((((((-------(.........)))))))))..).)).))....))))(-----(((.....)))).- ( -23.00, z-score = -0.98, R) >droYak2.chr2L 17580527 77 + 22324452 -----UGGAUACUUAACAAUUCAUAGUCAAGGC-------UGGACGGG-AAGCCUUGAUCUUAAAACGGGGUGGUAC-----GGGUUUUACCCGG- -----....((((...(.......(((((((((-------(.......-.)))))))).)).......)...))))(-----(((.....)))).- ( -22.94, z-score = -1.68, R) >droEre2.scaffold_4929 8910718 77 - 26641161 -----UGGAUACUUUACAAUUCAUAGUCAAGGC-------UGGCCGGG-AAGCCUUGAUCUUAAAACGGGGUGGUAC-----GGGUUUUACCCGA- -----....((((...(.......(((((((((-------(.......-.)))))))).)).......)...))))(-----(((.....)))).- ( -22.74, z-score = -0.95, R) >droAna3.scaffold_13266 16309117 82 - 19884421 -----UGAAUACUUAAUAAUUCAACGUCAAGGC-------UGGUCGGAGAAGCCUUGAUCUUAAAACAGGGAGGUGUACGUAGGGGUUUACCCG-- -----.(.((((((...........((((((((-------(.........)))))))))............)))))).)...(((.....))).-- ( -21.70, z-score = -1.12, R) >dp4.chr3 9634943 81 + 19779522 UCGUACGAAUACUUAAAAAUUCAACGUCAAGGC-------UGGUCAGGGGAGCCUUGAUCUUAAAACAGGGUGGCAC-----GGGGUUUAUCC--- ..((..((((........))))...((((((((-------(.........)))))))))......)).((((((...-----.....))))))--- ( -15.80, z-score = 0.50, R) >droPer1.super_4 4959620 81 + 7162766 UCGUACGAAUACUUAAAAAUUCAACGUCAAGGC-------UGGUCAGGGGAGCCUUGAUCUUAAAACAGGGUGGCAC-----GGGGUUUAUCC--- ..((..((((........))))...((((((((-------(.........)))))))))......)).((((((...-----.....))))))--- ( -15.80, z-score = 0.50, R) >droWil1.scaffold_181009 1282949 84 - 3585778 -----UAAAUGCUUAACAAUUCAACGUCAAGGCCCUAUGGUAGCAGAAAAAGCCUUGAUCUCAAAA-AGUAUGUUUU------UGUUUUCUUCUUU -----...((((((...........((((((((....((....))......))))))))......)-))))).....------............. ( -11.53, z-score = 0.20, R) >droVir3.scaffold_12875 20590082 80 - 20611582 -----UGAAUGUGCAGCGAUUCAACGUCAAGGC-------UGUGCCAAGGAGCCUUGAUCUCAACAGCCAACGGGCCG---AGGGAAACUCCCAA- -----(((((........)))))..((((((((-------(.(.....).))))))))).....(.(((....))).)---.(((.....)))..- ( -23.80, z-score = -0.35, R) >droGri2.scaffold_15245 10922588 78 + 18325388 -----UCAAUGUGCAUUGAUUCAACGUCAAGGC-------UAUGCUGA--AGCCUUGAUCUCAACAGUCAACGGGGCG---CGGGAAACACCCAA- -----.....((((.((((((....((((((((-------(.......--)))))))))......))))))....)))---)(((.....)))..- ( -25.00, z-score = -2.14, R) >consensus _____UGAAUACUUAACAAUUCAACGUCAAGGC_______UGGACGGGGAAGCCUUGAUCUUAAAACGGGGUGGUAC_____GGGUUUUACCCGG_ ......((((........))))...((((((((..................)))))))).......................(((.....)))... (-11.52 = -11.43 + -0.08)



| Location | 4,919,946 – 4,920,024 |
|---|---|
| Length | 78 |
| Sequences | 11 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 70.65 |
| Shannon entropy | 0.55782 |
| G+C content | 0.47581 |
| Mean single sequence MFE | -18.83 |
| Consensus MFE | -8.46 |
| Energy contribution | -8.64 |
| Covariance contribution | 0.18 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.839142 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 4919946 78 - 21146708 -CCGGGAAAAACCC-----GUACCACCCCGUUUUAAGAUCAAGGCUUCCCCGUCCA-------GCCUUGACUAUGAAUUGUUAAGUAUCCA----- -.((((.....)))-----)(((....(.((((..((.((((((((.........)-------)))))))))..)))).)....)))....----- ( -17.00, z-score = -1.74, R) >droSim1.chr2R 3589706 78 - 19596830 -CCGGGCAAAACCC-----GUACCACCCCGUUUUAAGAUCAAGGCUUCCCCGUCCA-------GCCUUGACUAUGAAUCGUUAAGUAUCCA----- -.((((.....)))-----)........((.((((((.((((((((.........)-------))))))))).)))).))...........----- ( -18.50, z-score = -2.32, R) >droSec1.super_1 2558733 78 - 14215200 -CCGGGAAAAACCC-----GUACCACCCCGUUUUAAGAUCAAGGCUUCCCCGUCCA-------GCCUUGACUAUGAAUCGUUAAGUAUCCA----- -.((((.....)))-----)........((.((((((.((((((((.........)-------))))))))).)))).))...........----- ( -18.50, z-score = -2.26, R) >droYak2.chr2L 17580527 77 - 22324452 -CCGGGUAAAACCC-----GUACCACCCCGUUUUAAGAUCAAGGCUU-CCCGUCCA-------GCCUUGACUAUGAAUUGUUAAGUAUCCA----- -.((((.....)))-----)(((....(.((((..((.((((((((.-.......)-------)))))))))..)))).)....)))....----- ( -18.60, z-score = -2.46, R) >droEre2.scaffold_4929 8910718 77 + 26641161 -UCGGGUAAAACCC-----GUACCACCCCGUUUUAAGAUCAAGGCUU-CCCGGCCA-------GCCUUGACUAUGAAUUGUAAAGUAUCCA----- -.((((.....)))-----)(((....(.((((..((.((((((((.-.......)-------)))))))))..)))).)....)))....----- ( -18.40, z-score = -1.46, R) >droAna3.scaffold_13266 16309117 82 + 19884421 --CGGGUAAACCCCUACGUACACCUCCCUGUUUUAAGAUCAAGGCUUCUCCGACCA-------GCCUUGACGUUGAAUUAUUAAGUAUUCA----- --.(((....)))..................(((((..((((((((.........)-------)))))))..)))))..............----- ( -16.60, z-score = -1.06, R) >dp4.chr3 9634943 81 - 19779522 ---GGAUAAACCCC-----GUGCCACCCUGUUUUAAGAUCAAGGCUCCCCUGACCA-------GCCUUGACGUUGAAUUUUUAAGUAUUCGUACGA ---((......))(-----((((...............((((((((.........)-------)))))))....((((........))))))))). ( -17.80, z-score = -2.11, R) >droPer1.super_4 4959620 81 - 7162766 ---GGAUAAACCCC-----GUGCCACCCUGUUUUAAGAUCAAGGCUCCCCUGACCA-------GCCUUGACGUUGAAUUUUUAAGUAUUCGUACGA ---((......))(-----((((...............((((((((.........)-------)))))))....((((........))))))))). ( -17.80, z-score = -2.11, R) >droWil1.scaffold_181009 1282949 84 + 3585778 AAAGAAGAAAACA------AAAACAUACU-UUUUGAGAUCAAGGCUUUUUCUGCUACCAUAGGGCCUUGACGUUGAAUUGUUAAGCAUUUA----- .............------.......((.-.((..(..((((((((((............))))))))))..)..))..))..........----- ( -15.40, z-score = -0.68, R) >droVir3.scaffold_12875 20590082 80 + 20611582 -UUGGGAGUUUCCCU---CGGCCCGUUGGCUGUUGAGAUCAAGGCUCCUUGGCACA-------GCCUUGACGUUGAAUCGCUGCACAUUCA----- -..(((.....))).---(((((....)))))(..(..((((((((.........)-------)))))))..)..)...............----- ( -22.00, z-score = 0.71, R) >droGri2.scaffold_15245 10922588 78 - 18325388 -UUGGGUGUUUCCCG---CGCCCCGUUGACUGUUGAGAUCAAGGCU--UCAGCAUA-------GCCUUGACGUUGAAUCAAUGCACAUUGA----- -..((((((.....)---)))))((((((...(..(..((((((((--.......)-------)))))))..)..).))))))........----- ( -26.50, z-score = -2.48, R) >consensus _CCGGGUAAAACCC_____GUACCACCCCGUUUUAAGAUCAAGGCUUCCCCGUCCA_______GCCUUGACGUUGAAUUGUUAAGUAUUCA_____ ...((......)).........................(((((((..................))))))).......................... ( -8.46 = -8.64 + 0.18)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:09:28 2011