
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 4,841,657 – 4,841,813 |
| Length | 156 |
| Max. P | 0.941450 |

| Location | 4,841,657 – 4,841,813 |
|---|---|
| Length | 156 |
| Sequences | 6 |
| Columns | 156 |
| Reading direction | reverse |
| Mean pairwise identity | 82.77 |
| Shannon entropy | 0.32402 |
| G+C content | 0.58451 |
| Mean single sequence MFE | -41.40 |
| Consensus MFE | -31.97 |
| Energy contribution | -34.17 |
| Covariance contribution | 2.20 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.941450 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 4841657 156 - 21146708 UCAACCACUCGCUGAUUGGCAUCUCCUGACCACAAAUUGGGAUUUGCCAUAACCAUGCCCCCGGGGCACCUGGCCGGGCACACGCUCGCCCAGACAGUCACAUAGACAUCCAUAGCCACUACCAUAUCCACAUCUACCGCCACCGACACCGCCGAG .......((((.((..(((((..(((..(.......)..)))..)))))............((((((..((((.(((((....))))).))))...(((.....)))...............................))).)))....)).)))) ( -44.60, z-score = -2.56, R) >droAna3.scaffold_13266 16230943 122 + 19884421 UCAACCGCCAGCUGAUUGGUAUCUCCU---------UUGGGG-CUGGCAUAACCAUACCCACGGGGCACCUGGCCGGGCACACGCUUACCCAGACAGCCACAGACACAUCCAUA---------AAGGCAGACUUUGCCGAA--------------- ......(((((((....((.....)).---------....))-))))).............((((((..((((..((((....))))..))))...))).............((---------(((.....))))))))..--------------- ( -34.50, z-score = 0.31, R) >droEre2.scaffold_4929 8832802 144 + 26641161 UCAACCACUUGCUGAUUGGCAUCUCCUGACCACAAAUUCGGAUUUGCCAUAACCAUGCCCCCGGGGCACCUGGCCGGGCACACGCUCGCCCAGACAGUCACAUAGACAUCCAC------------AUCCACAUCUACCGCCACCGACACCGCCGAG .......((((.((..(((((..(((.((........)))))..)))))............((((((..((((.(((((....))))).))))...(((.....)))......------------.............))).)))....)).)))) ( -40.40, z-score = -2.30, R) >droYak2.chr2L 17499651 144 - 22324452 UCAACCACUUGCUGAUUGGCAUCUCCUGACCACAAAUUGGGAUUUGCCAUAACCAUGCCCCCGAGGCACCUGGCCGGGCACACGCUCGCCCAGACAGUCACAUAGACAUCCAC------------AUCCACUUCUACCGCCAUCGACACCGCCGAG .......((((.((..(((((..(((..(.......)..)))..)))))............((((((..((((.(((((....))))).))))...(((.....)))......------------.............))).)))....)).)))) ( -40.40, z-score = -2.54, R) >droSec1.super_1 2479852 155 - 14215200 UCAACCACUCGCUGAUUGGCAUCUCCUGACCACAAAUUGGGAUUUGCCAUAACCAUG-CCCCGGGGCACCUGGCCGGGCACACGCUCGCCCAGACAGUCACAUAGACAUCCACAGCCACAUCCAUAUCCACAUCUACCGCCACCGCCACCGCCGAG .......((((.((..(((((..(((..(.......)..)))..)))))........-...((((((..((((.(((((....))))).))))...(((.....)))...............................))).)))....)).)))) ( -43.90, z-score = -2.12, R) >droSim1.chr2R 3511412 156 - 19596830 UCAACCACUCGCUGAUUGGCAUCUCCUGACCACAAAUUGGGAUUUGCCAUAACCAUGCCCCCGGGGCACCUGGCCGGGCACACGCUCGCCCAGACAGUCACAUAGACAUCCACAGCCACAUCCAUAUCCACAUCUACCGCCACCGACACCGCCGAG .......((((.((..(((((..(((..(.......)..)))..)))))............((((((..((((.(((((....))))).))))...(((.....)))...............................))).)))....)).)))) ( -44.60, z-score = -2.45, R) >consensus UCAACCACUCGCUGAUUGGCAUCUCCUGACCACAAAUUGGGAUUUGCCAUAACCAUGCCCCCGGGGCACCUGGCCGGGCACACGCUCGCCCAGACAGUCACAUAGACAUCCACA_________AUAUCCACAUCUACCGCCACCGACACCGCCGAG .......((((.((..(((((..((((((.......))))))..)))))............((((((..((((.(((((....))))).))))...(((.....)))...............................))).)))....)).)))) (-31.97 = -34.17 + 2.20)
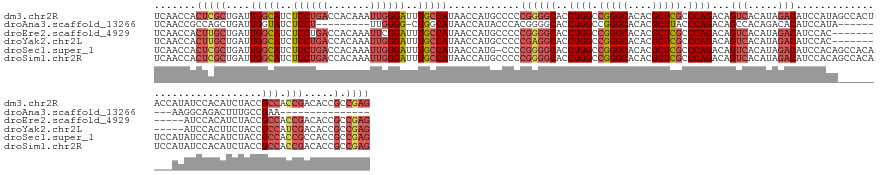
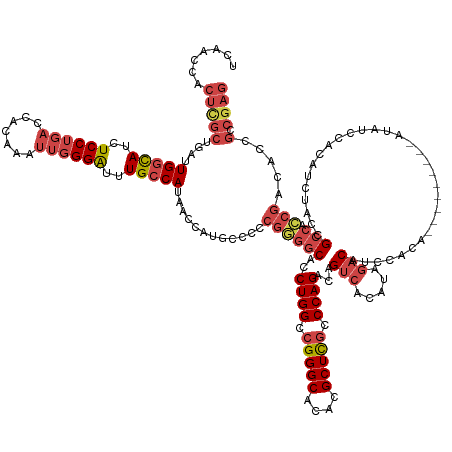

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:09:27 2011