| Sequence ID | dm3.chr2R |
|---|---|
| Location | 4,840,216 – 4,840,309 |
| Length | 93 |
| Max. P | 0.565969 |

| Location | 4,840,216 – 4,840,309 |
|---|---|
| Length | 93 |
| Sequences | 9 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 63.25 |
| Shannon entropy | 0.77914 |
| G+C content | 0.55881 |
| Mean single sequence MFE | -31.39 |
| Consensus MFE | -7.27 |
| Energy contribution | -7.89 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.23 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.565969 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 4840216 93 - 21146708 ACGAGGCGGGGCCGCAGGAGCAGGAGGAGC---GGCAGGAG--GACACC-UCAGACAGCCACUAAUUGGCCCCGAACUGCGCAUUUAUCGCGUAGGACA ......(((((((((....))..((..((.---(((..(((--(...))-)).....))).))..)))))))))..((((((.......)))))).... ( -37.60, z-score = -2.60, R) >droEre2.scaffold_4929 8831380 70 + 26641161 -----------------------AAGAGGC---GGCAAAAG--GACACC-UCAGACAGCCACUAAUUGGCCCCGAACUGCGCAUUUAUCGCGUAGGACA -----------------------..((((.---(.......--..).))-)).....((((.....))))...(..((((((.......))))))..). ( -20.00, z-score = -1.37, R) >droSec1.super_1 2478430 93 - 14215200 ACGAGGCGGGGCCGCAGGAGCAGAAGGAGC---GGCAAGAG--GACACC-UCAGACAGCCACUAAUUGGCCCCGAACUGCGCAUUUAUCGCGUAGGACA ......(((((((((....))..((..((.---(((..(((--(...))-)).....))).))..)))))))))..((((((.......)))))).... ( -36.10, z-score = -2.61, R) >droSim1.chr2R 3509971 93 - 19596830 ACGAGGCGGGGCCGCAGGAGCAGGAGGAGC---GGCAAGAG--GACACC-UCAGACAGCCACUAAUUGGCCCCGAACUGCGCAUUUAUCGCGUAGGACA ......(((((((((....))..((..((.---(((..(((--(...))-)).....))).))..)))))))))..((((((.......)))))).... ( -36.50, z-score = -2.49, R) >dp4.chr3 8457486 97 + 19779522 UUGGGCCGUGCCCGUGCGAGUGGGGAGGGCCGGAGGAAGAA--GUCCUCUGCAGACGGCCUCUAAUUUCCCCUGUACUGCGCAUUUAUCGCGUAGGACU ..((((...))))........((((((((((((((((....--.)))))......))))))......))))).((.((((((.......)))))).)). ( -38.40, z-score = -0.60, R) >droPer1.super_4 437370 97 + 7162766 UUGGGCCGUGCCCGUGCGAGUGGGGAGGGCCGGAGGAAGAA--GUCCUCUGCAGACGGCCUCUAAUUUCCCCUGUACUGCGCAUUUAUCGCGUAGGACU ..((((...))))........((((((((((((((((....--.)))))......))))))......))))).((.((((((.......)))))).)). ( -38.40, z-score = -0.60, R) >droAna3.scaffold_13266 16229530 94 + 19884421 ACGAGCAGAACCUGCAACAGGAGGUGGAGG---AAGCAGAG--GACCUCGCCAGACAGCCACUAAUUGGCCCUGAAUUGCGCAUUUAUCGCGUACGAAU .((.((((...))))..((((.((((.(((---........--..))))))).....((((.....))))))))...(((((.......)))))))... ( -27.50, z-score = -1.12, R) >droVir3.scaffold_12875 20492675 98 + 20611582 ACAAAAC-UAUGUAAAGGAGACGGCAGCGCAGCAGUCAACGUCGACUGCAGACGGCGGCAACUAAUUUGCUUUGUAUUGCGCAUUUAUUGUAUAGGACA ......(-(((((((.(....)....(((((((((((......))))))..((((.(((((.....)))))))))..))))).....)))))))).... ( -29.40, z-score = -1.66, R) >droMoj3.scaffold_6496 5383065 73 - 26866924 ACAAAAC-UAUGCAAAGGAUACGGCAACGC-------------------------CGACAACUAAUUUGCUUUGUAUUGCGCAUUUAUUGCAUAGGACA ......(-(((((((..(((.((((...))-------------------------))...........((........))..)))..)))))))).... ( -18.60, z-score = -2.52, R) >consensus ACGAGCCGUGCCCGCAGGAGCAGGAGGAGC___AGCAAGAG__GACACC_UCAGACAGCCACUAAUUGGCCCUGAACUGCGCAUUUAUCGCGUAGGACA .........................................................((((.....))))......((((((.......)))))).... ( -7.27 = -7.89 + 0.62)


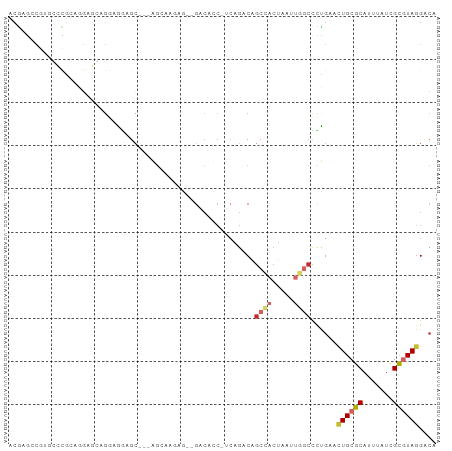
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:09:25 2011