
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 4,828,356 – 4,828,455 |
| Length | 99 |
| Max. P | 0.807501 |

| Location | 4,828,356 – 4,828,455 |
|---|---|
| Length | 99 |
| Sequences | 9 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 70.56 |
| Shannon entropy | 0.58077 |
| G+C content | 0.51918 |
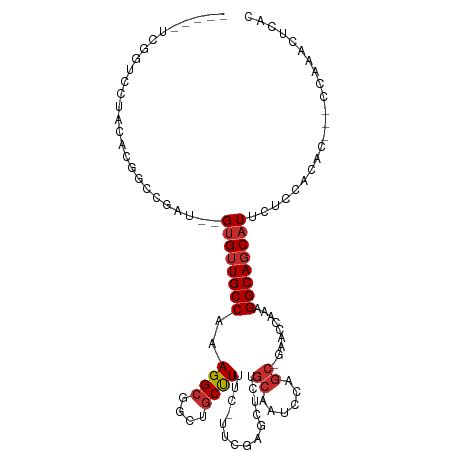
| Mean single sequence MFE | -26.71 |
| Consensus MFE | -13.44 |
| Energy contribution | -13.81 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.807501 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
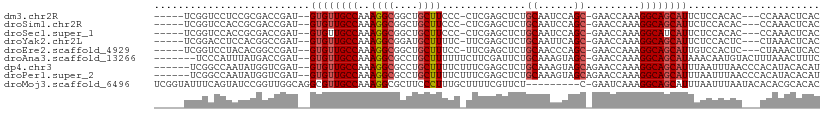
>dm3.chr2R 4828356 99 + 21146708 -----UCGGUCCUCCGCGACCGAU--GUGUUGCCAAAGGCGGCUGCUUCCC-CUCGAGCUCUGCAAUCCAGC-GAACCAAAGGCAGCAUUCUCCACAC---CCAAACUCAC -----((((((......)))))).--((((((((....((((..((((...-...)))).))))..((....-))......)))))))).........---.......... ( -27.70, z-score = -1.18, R) >droSim1.chr2R 3497817 99 + 19596830 -----UCGGUCCACCGCGACCGAU--GUGUUGCCAAAGGCGGCUGCUUCCC-CUCGAGCUCUGCAAUCCAGC-GAACCAAAGGCAGCAUUCUCCACAC---CCAAACUCAC -----((((((......)))))).--((((((((....((((..((((...-...)))).))))..((....-))......)))))))).........---.......... ( -27.70, z-score = -1.20, R) >droSec1.super_1 2466358 99 + 14215200 -----UCGGUCCACCGCGACCGAU--GUGUUGCCAAAGGCGGCUGCUUCCC-CUCGAGCUCUGCAAUCCAGC-GAACCAAAGGCAUCAUUCUCCACAC---CCAAACUCAC -----((((((......)))))).--(((.((((....((((..((((...-...)))).))))..((....-))......)))).))).........---.......... ( -23.60, z-score = -0.32, R) >droYak2.chr2L 17486175 99 + 22324452 -----UCGGACCUCCACGGCCGAU--GUGUUGCCAAAGGCGGAUGCUUUUC-UUCGAGCUCUGCAAUUCAGC-GAACCAAAGGCAGCAUUCUCCACUC---CUAAACUCAC -----((((.((.....)))))).--((((((((....(((((.((((...-...)))))))))..(((...-))).....)))))))).........---.......... ( -28.10, z-score = -1.29, R) >droEre2.scaffold_4929 8819515 99 - 26641161 -----UCGGUCCUACACGGCCGAU--GUGUUGCCAAAGGCGGCUGCUUUCC-UUCGAGCUCUGCAACCCAGC-GAACCAAAGGCAGCAUUGUCCACUC---CUAAACUCAC -----((((((......)))))).--(((.((((...))))((((((((..-((((....(((.....))))-)))...))))))))......)))..---.......... ( -26.40, z-score = -0.19, R) >droAna3.scaffold_13266 16218734 101 - 19884421 -------UCCCAUUUAUGACCGAU--GUGUUGCCAAAGGCGCCUGCUUUUUUCUUCGAUUCUGCAAAGUAGC-GAACCAAAGGCAGCAUAAACAAUGUACUUUAAACUUUC -------................(--((((((((((((((....))))))...((((...((....))...)-))).....)))))))))..................... ( -21.10, z-score = -0.80, R) >dp4.chr3 19167184 103 - 19779522 ------UCGGCCAAUAUGGUCGAU--GUGUUGCCAAAGGCGCCUGCUUUUCUUUCGAGCUCUGCAAAGUAGCAGAACCAAAGGCAGCAUUUAAUUUAACCCACAUACACAU ------((((((.....))))))(--((((.(((...)))((.(((((((..(((..(((((....)).))).)))..)))))))))..................))))). ( -30.80, z-score = -2.45, R) >droPer1.super_2 8257035 103 - 9036312 ------UCGGCCAAUAUGGUCGAU--GUGUUGCCAAAGGCGCCUGCUUUUCUUUCGAGCUCUGCAAAGUAGCAGAACCAAAGGCAGCAUUUAAUUUAACCCACAUACACAU ------((((((.....))))))(--((((.(((...)))((.(((((((..(((..(((((....)).))).)))..)))))))))..................))))). ( -30.80, z-score = -2.45, R) >droMoj3.scaffold_6496 5365967 101 + 26866924 UCGGUAUUUCAGUAUCCGGUUGGCAGGCGUUGCCAAAGGCGCUUCCCUUUGCUUUUCGUUCU---------C-GAAUCAAAGGCAGCAUUUAAUUUAAUACACACGCACAC ...(((((.......((..(((((((...))))))).)).(((..((((((...((((....---------)-))).)))))).))).........))))).......... ( -24.20, z-score = -1.18, R) >consensus _____UCGGUCCUACACGGCCGAU__GUGUUGCCAAAGGCGGCUGCUUUUC_UUCGAGCUCUGCAAUCCAGC_GAACCAAAGGCAGCAUUCUCCACAC___CCAAACUCAC ..........................((((((((..((((....))))...........((.((......)).))......))))))))...................... (-13.44 = -13.81 + 0.37)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:09:23 2011