
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 4,798,926 – 4,799,046 |
| Length | 120 |
| Max. P | 0.554175 |

| Location | 4,798,926 – 4,799,046 |
|---|---|
| Length | 120 |
| Sequences | 14 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.08 |
| Shannon entropy | 0.57496 |
| G+C content | 0.51432 |
| Mean single sequence MFE | -39.41 |
| Consensus MFE | -12.90 |
| Energy contribution | -12.35 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.71 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.554175 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 4798926 120 + 21146708 GUACCAAUCAACGCGCCUAGGGUGAAAGGUGAAACCUUCUUCACCACAAUCGAGUUUGUGGGCCGGUUGCCGACGAACACCUUGUGGGGCAACAAGUUAUCCAGCUCGUUGCCGCACAGG .............((.....(((((((((.....)))..)))))).....)).((((((.(((.....))).))))))..((((((.((((((.((((....)))).)))))).)))))) ( -45.70, z-score = -2.43, R) >apiMel3.Group1 6495093 120 + 25854376 UUACCAAUCAAAGCUCCAAGAAUAAAAGGCGUUAUUUUUUUAAGAAGGAUAGUAUUGGUUGGUCUGUUUCCUUCAAACACUUUAUGUGGUAAUAACAAAUUUAUUUGUUCAGGAUUUACG ..(((((((((.(((((((((((((......)))))))).......)))..)).)))))))))..((.((((.....(((.....))).....((((((....)))))).))))...)). ( -22.51, z-score = -0.84, R) >anoGam1.chr3L 3703308 114 - 41284009 ------AUCAGUGCGCCGAGCACGAACGGUGUCACCUUCUUCACCAGGAUCGAGUUCGUUGGCCGGUUGCCGGUGAACACCUUGUGCGGGAGCAGCUUCUCGAGCGCCUCACCCGCCAGU ------....((((.....))))....(((((((((..(...(((.((..((....))....))))).)..)))).)))))(((.(((((.((.((((...))))))....)))))))). ( -39.00, z-score = 0.59, R) >droGri2.scaffold_15112 3877561 120 + 5172618 UCACCAAUCAAGGCGCCUAAAACGAAGGGUGAAACCUUUUUGACUACAAUCGAAUUUGUUGGCCGAUUGCCGGCAAAGACUUUGUGGGGCAACAAGUUGUCCAGCUCGUUGCCGCACAAG ....(((..(((((((((........)))))...)))).)))............(((((((((.....)))))))))....(((((.((((((.((((....)))).)))))).))))). ( -40.40, z-score = -2.28, R) >droMoj3.scaffold_6496 13157814 120 - 26866924 UUACCAAUCAAUGCACCUAAAGUGAAUGGUGAGACUUUCUUGACCACAAUCGAGUUGGUCGGGCGGUUGCCAACGAAGACCUUAUGGGGCAACAAGUUUUCCAGCUCGUUGCCGCACAGG ((((((.(((.(........).))).))))))((((..(((((......)))))..)))).(((....)))........(((..((.((((((.(((......))).)))))).)).))) ( -37.50, z-score = -1.29, R) >droVir3.scaffold_12875 1782044 120 + 20611582 UUACCAAUCAAAGCACCCAAAGUGAAUGGUGACACCUUCUUGACUACAAUCGAGUUGGUCGGCCUGUUGCCGGCAAACACCUUGUGUGGCAACAAGUUGUCCAGCUCGUUGCCACACAAG ((((((.(((............))).))))))...(..(((((......)))))..)((((((.....))))))......(((((((((((((.((((....)))).))))))))))))) ( -44.80, z-score = -3.72, R) >droWil1.scaffold_180700 1347391 120 - 6630534 UCACCUAUCAAAGCGCCCAAAGUGAACGGCGACACCUUCUUGACGACAAUUGAAUUUGUUGGACGAUUGCCAACAAAUACCUUGUGGGGCAAGAGGUUCUCCAGCUCAUUGCCACACACA .....................(((...((.((.((((.((((.(.((((..(.(((((((((.......))))))))).).)))).)..)))))))))).)).((.....))..)))... ( -30.70, z-score = -0.84, R) >droPer1.super_2 8020775 120 + 9036312 UUACCAAUCAAUGCGCCCAGUGUAAAUGGCGACACUUUCUUCACCACAAUUGAGUUCGUCGGCCGAUUGCCAGCGAACACUUUGUGCGGCAGCAGGUUUUCCAGCUCGUUGCCGCACAAU .......(((((((((...))))...(((.((........)).)))..)))))((((((.(((.....))).))))))...((((((((((((..((......))..)))))))))))). ( -44.40, z-score = -3.75, R) >dp4.chr3 6180456 120 - 19779522 GUACCAAUCAAUGCGCCCAGUGUAAAUGGCGACACUUUCUUCACCACAAUUGAGUUCGUCGGCCGAUUGCCAGCGAACACUUUGUGCGGCAGCAGGUUUUCCAGCUCGUUGCCGCACAAU .......(((((((((...))))...(((.((........)).)))..)))))((((((.(((.....))).))))))...((((((((((((..((......))..)))))))))))). ( -44.40, z-score = -3.53, R) >droAna3.scaffold_13266 18666028 120 - 19884421 UCACCAAUCAACGCGCCCAAUGUGAACGGGGAUACCUUCUUCACAACAAUUGAGUUCGUCGGGCGAUUGCCAACGAAUACUUUGUGAGGCAGCAAGUUAUCCAGUUCAUUGCCGCACAAG ............(((.(....((((((..(((((.(((((((((((.....(.((((((..(((....))).)))))).).))))))))....))).))))).)))))).).)))..... ( -35.00, z-score = -1.92, R) >droEre2.scaffold_4929 8790100 120 - 26641161 GUACCAAUCAACGCGCCCAGGGUGAAAGGUGAGACCUUCUUCACCACAAUAGAGUUUGUGGGCCGGUUGCCAACGAACACCUUGUGGGGCAACAAGUUAUCCAGCUCGUUGCCGCACAGA ............((.((((((((....((((((......))))))........((((((.(((.....))).)))))))))))).))((((((.((((....)))).))))))))..... ( -44.00, z-score = -2.09, R) >droYak2.chr2L 17456953 120 + 22324452 GUACCAAUCAACGCGCCCAGGGUGAAAGGUGAGACCUUCUUCACCACAAUCGAGUUUGUGGGCCGGUUGCCAACGAACACCUUGUGGGGCAACAAGUUGUCCAGCUCGUUGCCGCACAGA ............((.((((((((....((((((......))))))........((((((.(((.....))).)))))))))))).))((((((.((((....)))).))))))))..... ( -44.90, z-score = -1.65, R) >droSec1.super_1 2436791 120 + 14215200 GUACCAAUCAACGCGCCCAGGGUGAAAGGUGAGACCUUCUUCACCACAAUCGAGUUUGUGGGCCGGUUGCCGACGAACACCUUGUGGGGCAACAAGUUAUCCAGCUCGUUGCCGCACAGA ............((.((((((((....((((((......))))))........((((((.(((.....))).)))))))))))).))((((((.((((....)))).))))))))..... ( -46.10, z-score = -2.26, R) >triCas2.ChLG7 14975702 114 - 17478683 ------AUGAGCGCGCCGAGCGUAAACGGCGUGAUUUUCUUCACCAUAAUACUGUUAGUCGGCCUGUUCCCUUCGAAGAUCUUAUGCGGGAGGAUCUUCUCCAGGGCCUCUCCCCCCAGC ------..(((((.((((((((((....(.((((......)))))....))).))...))))).))))).....(((((((((.......)))))))))....(((........)))... ( -32.30, z-score = 0.23, R) >consensus UUACCAAUCAACGCGCCCAGAGUGAAAGGUGACACCUUCUUCACCACAAUCGAGUUUGUCGGCCGGUUGCCAACGAACACCUUGUGGGGCAACAAGUUAUCCAGCUCGUUGCCGCACAGG .............((((..........)))).............(((((.....(((((.((.......)).)))))....))))).(((.(..((((....))))..).)))....... (-12.90 = -12.35 + -0.55)

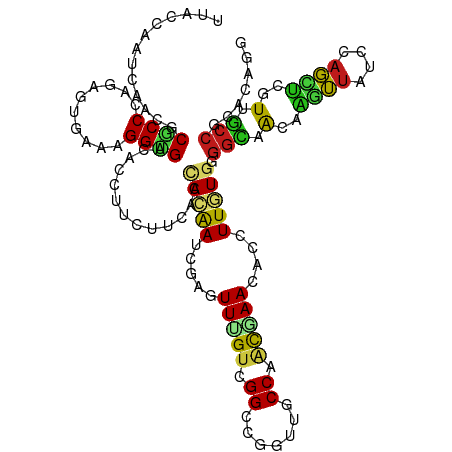
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:09:21 2011