| Sequence ID | dm3.chr2L |
|---|---|
| Location | 2,392,598 – 2,392,705 |
| Length | 107 |
| Max. P | 0.861139 |

| Location | 2,392,598 – 2,392,705 |
|---|---|
| Length | 107 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 79.44 |
| Shannon entropy | 0.44644 |
| G+C content | 0.43415 |
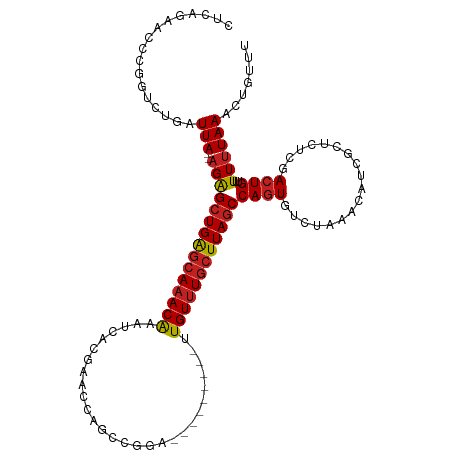
| Mean single sequence MFE | -29.70 |
| Consensus MFE | -14.31 |
| Energy contribution | -14.76 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.861139 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
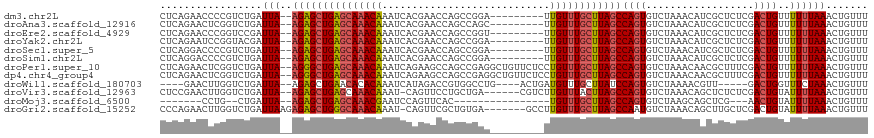
>dm3.chr2L 2392598 107 - 23011544 CUCAGAACCCCGUCUGAUUA--AGAGCUGAGCAAACAAAUCACGAACCAGCCGGA---------UUGUUUGCUUAGCCAGUGUCUAAACAUCGCUCUCGACUGUUUUUUAAACUGUUU .(((((......)))))...--...(((((((((((((.((.((.......))))---------)))))))))))))((((....((((((((....))).))))).....))))... ( -32.60, z-score = -3.92, R) >droAna3.scaffold_12916 9554827 107 - 16180835 CUCAGAACUCGGUCUGAUUA--AGAGCUGAGCAAACAAAUCACGAACCAGCCAGC---------UUGUUUGCUUAGCCAGUGUCUAAACAUCGCUCUCGACUGUUUUUUAAACUGUUU .(((((......)))))...--...(((((((((((((.................---------)))))))))))))((((....((((((((....))).))))).....))))... ( -28.83, z-score = -1.90, R) >droEre2.scaffold_4929 2434547 107 - 26641161 CUCAGAACCCGGUCCGAUUA--AGAGCUGAGCAAACAAAUCACGAACCAGCCGGU---------UUGUUUGCUUAGCCAGUGUCUAAACAUCGCUCUCGACUGUUUUUUAAACUGUUU ...((((..(((((.((...--.(.((((((((((((((((.(......)..)))---------))))))))))))))((((.........)))).)))))))..))))......... ( -31.10, z-score = -2.87, R) >droYak2.chr2L 2377443 107 - 22324452 CUCAGAAUCCGGUACGAUUA--AGAGCUGAGCAAACAAAUCACGAACCAGCCGGA---------UUGUUUGCUUAGCCAGUGUCUAAACAUCGCUCUCGACUGUUUUUUAAACUGUUU ...((((..((((.(((...--.(.(((((((((((((.((.((.......))))---------))))))))))))))((((.........)))).)))))))..))))......... ( -30.20, z-score = -2.62, R) >droSec1.super_5 566034 107 - 5866729 CUCAGGACCCCGUCUGAUUA--AGAGCUGAGCAAACAAAUCACGAACCAGCCGGA---------UUGUUUGCUUAGCCAGUGUCUAAACAUCGCUCUCGACUGUUUUUUAAACUGUUU .(((((......)))))...--...(((((((((((((.((.((.......))))---------)))))))))))))((((....((((((((....))).))))).....))))... ( -31.70, z-score = -3.06, R) >droSim1.chr2L 2351691 107 - 22036055 CUCAGGACCCCGUCUGAUUA--AGAGCUGAGCAAACAAAUCACGAACCAGCCGGA---------UUGUUUGCUUAGCCAGUGUCUAAACAUCGCUCUCGACUGUUUUUUAAACUGUUU .(((((......)))))...--...(((((((((((((.((.((.......))))---------)))))))))))))((((....((((((((....))).))))).....))))... ( -31.70, z-score = -3.06, R) >droPer1.super_10 654245 116 - 3432795 CUCAGAACUCGGUCUGAUUA--AGGGCUGAGCAAACAAAUCAGAAGCCAGCCGAGGCUGUUCUCCUGUUUGCUUAGCCAGUGUCUAAACAACGCUUUCGACUGUUUUUUAAACUGUUU ...((((..(((((......--..(((((((((((((....((((..((((....))))))))..)))))))))))))(((((.......)))))...)))))..))))......... ( -39.00, z-score = -3.03, R) >dp4.chr4_group4 1650545 116 - 6586962 CUCAGAACUCGGUCUGAUUA--AGGGCUGAGCAAACAAAUCAGAAGCCAGCCGAGGCUGUUCUCCUGUUUGCUUAGCCAGUGUCUAAACAACGCUUUCGACUGUUUUUUAAACUGUUU ...((((..(((((......--..(((((((((((((....((((..((((....))))))))..)))))))))))))(((((.......)))))...)))))..))))......... ( -39.00, z-score = -3.03, R) >droWil1.scaffold_180703 1057435 103 + 3946847 ----GAACUUGGUCUGAUUA--AGAGCUGAACACACAAAUCAUAGACCGUGGCCUG----ACUGAUGUUUGCUUAUCCAGUGUCUAAAACGUU-----GACUGGUUUCUAAACUGUUU ----(((.(..(((......--.((..((......))..)).((((((..(((..(----((....))).)))......).))))).......-----)))..).))).......... ( -16.40, z-score = 1.41, R) >droVir3.scaffold_12963 9725490 109 - 20206255 CUCCGAACUUGGUCUGAUUA--AGAGCUGAGCAAACAAAU-CAGUUCCUGCUGA------CGUCUUGUUUACUUAGCCAGUGUCUAAACAGCUCUCUCGACUGUAUUUUAAACUGUUU .......((((((...))))--)).((((((.((((((.(-((((....)))))------....)))))).))))))((((......((((.((....)))))).......))))... ( -24.42, z-score = -0.85, R) >droMoj3.scaffold_6500 30821329 88 + 32352404 -------CCUG--CUGAUUA--AGAGCUGAGCAAACGAAUCCAGUUCAC----------------UGUUUGCUUAGCCAGUGUCUAAGCAGCUCG---AACUGUAUUUUAAACUGUUU -------.(((--(((((..--.(.(((((((((((((((...))))..----------------.))))))))))))...)))..)))))...(---(((.((.......)).)))) ( -23.70, z-score = -1.67, R) >droGri2.scaffold_15252 5113902 110 + 17193109 CCCAGAACUUGGUCUGAUUAAGAGAGCUGGGCAAACAAAU-CAGUUCGCUGUGA-------GCCUUGUUUGCUUAGCCAAUGUCUAAACAGCUUGCUCGACUGUAUUUUAAACUGUUU ...((((..(((((.((.((((.(.(((((((((((((..-..((((.....))-------)).))))))))))))))..(((....))).)))).)))))))..))))......... ( -27.80, z-score = -0.53, R) >consensus CUCAGAACCCGGUCUGAUUA__AGAGCUGAGCAAACAAAUCACGAACCAGCCGGA_________UUGUUUGCUUAGCCAGUGUCUAAACAUCGCUCUCGACUGUUUUUUAAACUGUUU .........................((((((((((((............................))))))))))))((((......(((...........))).......))))... (-14.31 = -14.76 + 0.45)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:11:07 2011