
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 4,795,913 – 4,796,023 |
| Length | 110 |
| Max. P | 0.513547 |

| Location | 4,795,913 – 4,796,023 |
|---|---|
| Length | 110 |
| Sequences | 11 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 65.71 |
| Shannon entropy | 0.74680 |
| G+C content | 0.41451 |
| Mean single sequence MFE | -29.11 |
| Consensus MFE | -8.58 |
| Energy contribution | -7.84 |
| Covariance contribution | -0.74 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.29 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.513547 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
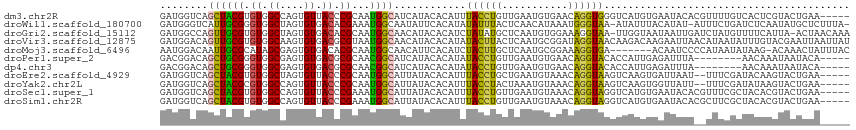
>dm3.chr2R 4795913 110 + 21146708 GAUGGUCAGCUACGUGUGGCCAGUGUUACCCGCAAUGGCAUCAUACACAUUUACCUGUUGAAUGUGAACAGGUGGGUCAUGUGAAUACACGUUUUGUCACUCGUACUGAA----- .....((((.((((.(((((..((((....((((.((((.((((.((((((((.....)))))))).....))))))))))))...)))).....))))).)))))))).----- ( -35.60, z-score = -1.03, R) >droWil1.scaffold_180700 1344438 112 - 6630534 GAUGGGUCAUUGCGGGUGGCUAGUGUGACACGAAAUGGCAAUAUUCACAUAUAUUUACUCAACAUAAAUGGGUAA-AUAUUUACAUAU-AUUUCUGAUCUCAAUAUGCUCUUUA- (((.(((((((.((.((.((....)).)).)).))))))(((((......(((((((((((.......)))))))-))))......))-)))..).)))...............- ( -23.60, z-score = -1.39, R) >droGri2.scaffold_15112 3874238 113 + 5172618 GAUGGCCAGUUGCGUGUGGCUAGUGUGACACGCAAUGGCAACAUACACAUCUAUAUGCUCAAUGUGGAAAGGUAA-UUGGUAAUAAUUGAUCUAUGUUUUCAUUA-ACUAACAAA ((((((((.((((((((.((....)).))))))))))))(((((.....(((((((.....))))))).((((((-(((....))))).)))))))))..)))).-......... ( -33.50, z-score = -2.43, R) >droVir3.scaffold_12875 1778903 115 + 20611582 GAUGGACAGUUGCGUGUGGCAAGUGUGACGCGUAAUGGCAACAUACACAUAUACUUACUCAAUGCGGAUAGGUAACAAGACAAGAAUUAACAUAAUAUUUGUACGAAUUAAUUAU .(((....(((((((((.((....)).)))))))))(....).....))).((((((..(.....)..)))))).....(((((.((((...)))).)))))............. ( -22.60, z-score = -0.15, R) >droMoj3.scaffold_6496 13153942 107 - 26866924 AAUGGACAAUUGCGCAUAGCGAGUGUGACACGCAAUGGCAACAUUCACAUCUACUUGCUCAAUGCGGAAAGGUGA-------ACAAUCCCCAUAAUAUAAG-ACAAACUAUUUAC .((((....((.(((((((((((((.((...(.((((....)))))...))))))))))..))))).)).((...-------.....))))))........-............. ( -25.20, z-score = -1.88, R) >droPer1.super_2 8017687 102 + 9036312 GACGGACAGCUGCGGGUGGCGAGUGUGACGCGCAACGGCAUCAUACACAUAUACCUGUUGAAUGUGAACAGGUACACCAUUGAGAUUUA--------AACAAAUAAUACA----- (.(((....)))).((((....((((((.((......)).)))))).....((((((((.......))))))))))))...........--------.............----- ( -25.70, z-score = -0.84, R) >dp4.chr3 6177376 102 - 19779522 GACGGACAGCUGCGGGUGGCGAGUGUGACGCGCAACGGCAUCAUACACAUAUACCUGUUGAAUGUGAACAGGUACACCAUUGAGAUUUA--------AACAAAUAAUACA----- (.(((....)))).((((....((((((.((......)).)))))).....((((((((.......))))))))))))...........--------.............----- ( -25.70, z-score = -0.84, R) >droEre2.scaffold_4929 8787077 108 - 26641161 GAUGGUCAGCUACGUGUGGCUAGUGUUACCCGCAAUGGCAUUAUACACAUUUACCUGCUGAAUGUAAACAGGUAAGUCAAGUGAUUAAU--UUUCGAUACAAGUACUGAA----- .....((((.(((.((((((((.(((.....))).))))......((((((((((((...........)))))))))...)))......--......)))).))))))).----- ( -25.40, z-score = -0.26, R) >droYak2.chr2L 17453920 108 + 22324452 GAUGGUCAGCUACGCGUGGCCAGUGUUACCCGCAAUGGCAUUAUACACAUUUACCUACUAAAUGUAAACAGGUAAGUCAAGUGGUUAUU--UUUCGAUAUAAGUACUGAA----- ..((((((((...)).))))))((((.....((....)).....))))((((((((((.....))....))))))))..((((.((((.--.......)))).))))...----- ( -23.60, z-score = 0.12, R) >droSec1.super_1 2433738 110 + 14215200 GAUGGUCAGCUACGUGUGGCCAGUGUUACCCGAAAUGGCAUUAUACACAUUUACCUGUUGAAUGUAAACAGGUAGGUCAUGUGAAUACACGUUUCGCUACACGUACUGAA----- .....((((.((((((((((..(.....)..((((((....(((.((((((((((((((.......)))))))))...))))).)))..)))))))))))))))))))).----- ( -39.40, z-score = -3.25, R) >droSim1.chr2R 3469182 110 + 19596830 GAUGGUCAGCUACGUGUGGCCAGUGUUACCCGAAAUGGCAUUAUACACAUUUACCUGUUGAAUGUAAACAGGUAGGUCAUGUGAAUACACGCUUCGCUACACGUACUGAA----- .....((((.((((((((((.(((((..((......)).......((((((((((((((.......)))))))))...))))).....)))))..)))))))))))))).----- ( -39.90, z-score = -3.37, R) >consensus GAUGGUCAGCUGCGUGUGGCCAGUGUGACCCGCAAUGGCAUCAUACACAUUUACCUGCUGAAUGUGAACAGGUAAGUCAUGUAAAUAUA__UUUCGAUACAAAUACUGAA_____ ........((((((.((.((....)).)).))...))))............((((((...........))))))......................................... ( -8.58 = -7.84 + -0.74)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:09:20 2011