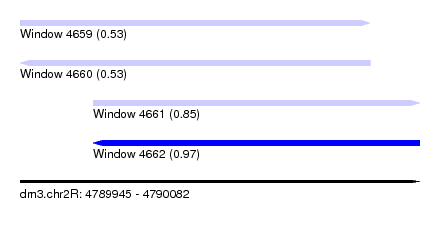
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 4,789,945 – 4,790,082 |
| Length | 137 |
| Max. P | 0.973627 |

| Location | 4,789,945 – 4,790,065 |
|---|---|
| Length | 120 |
| Sequences | 15 |
| Columns | 121 |
| Reading direction | forward |
| Mean pairwise identity | 78.71 |
| Shannon entropy | 0.48956 |
| G+C content | 0.49389 |
| Mean single sequence MFE | -41.25 |
| Consensus MFE | -8.17 |
| Energy contribution | -7.32 |
| Covariance contribution | -0.85 |
| Combinations/Pair | 1.72 |
| Mean z-score | -2.98 |
| Structure conservation index | 0.20 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.534366 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
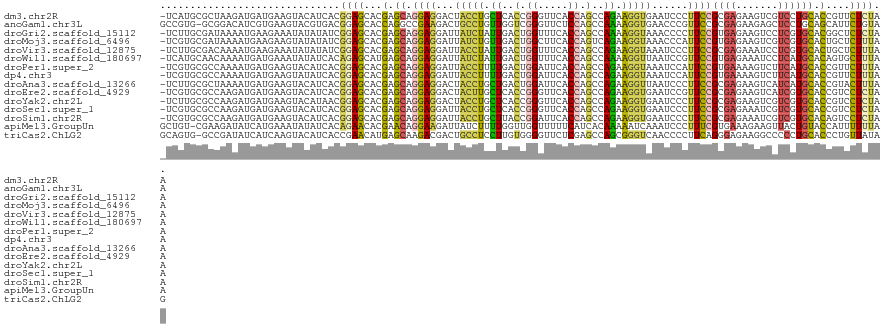
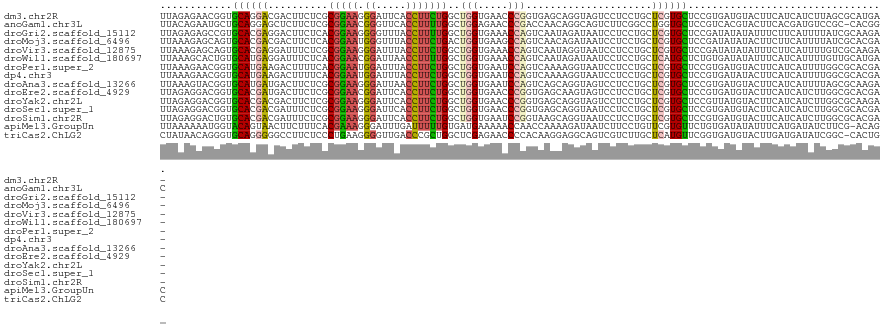
>dm3.chr2R 4789945 120 + 21146708 -UCAUGCGCUAAGAUGAUGAAGUACAUCACGGAGCACGAGCAGGAGGACUACCUGCUCACCGGGUUCACCAGCCAGAAGGUGAAUCCCUUCCGCGAGAAGUCGUCCUGCACCGUUCUCUAA -.............(((((.....))))).((((.(((.((((((.((((.(.(((.....(((((((((........))))))))).....))).).)))).))))))..))).)))).. ( -46.00, z-score = -3.13, R) >anoGam1.chr3L 5195294 120 + 41284009 GCCGUG-GCGGACAUCGUGAAGUACGUGACGGAGCACCAGGCCGAAGACUGCCUGUUGGUCGGGUUCUCCAGCCAAAAGGUGAACCCGUUCCGCGAGAAGAGCUCCUGCAGCAUUCUGUAA ...((.-(((.((........)).))).))(((((..(((((........)))))..((.(((((((.((........)).)))))))..)).........)))))(((((....))))). ( -44.30, z-score = -0.96, R) >droGri2.scaffold_15112 3867477 120 + 5172618 -UCUUGCGAUAAAAUGAAGAAAUAUAUAUCGGAGCACGAGCAGGAGGAUUAUCUAUUGACUGGUUUCACCAGCCAAAAGGUAAACCCCUUCCGUGAGAAGUCCUCGUGCACGGCUCUCUAA -.....(((((..((......))...)))))((((.((.(((.(((((((((((.(((.((((.....)))).))).))))))....(((.(....))))))))).))).))))))..... ( -39.70, z-score = -3.47, R) >droMoj3.scaffold_6496 13147778 120 - 26866924 -UCGUGCGAUAAAAUGAAGAAGUAUAUAUCGGAGCACGAGCAGGAGGAUUAUCUGUUGACUGGCUUCACCAGUCAGAAGGUAAACCCAUUCCGUGAGAAGUCGUCGUGCACUGCUCUUUAA -((((........)))).............((((((.(.(((.((.((((((((.((((((((.....)))))))).))))).........(....)..))).)).))).))))))).... ( -36.60, z-score = -1.87, R) >droVir3.scaffold_12875 1773228 120 + 20611582 -UCUUGCGACAAAAUGAAGAAAUAUAUAUCGGAGCACGAGCAGGAGGAUUACCUAUUGACUGGUUUCACCAGCCAGAAGGUAAAUCCCUUCCGCGAGAAAUCCUCGUGCACUGCUCUUUAA -...(((..(...(((.(....).)))...)..))).((((((..(((((((((.(((.((((.....)))).))).)))).)))))....((((((.....))))))..))))))..... ( -39.00, z-score = -3.63, R) >droWil1.scaffold_180697 1470382 120 - 4168966 -UCAUGCAACAAAAUGAUGAAAUAUAUCACAGAGCAUGAGCAGGAGGAUUAUCUAUUGACUGGUUUCACCAGCCAAAAGGUUAAUCCGUUCCGUGAGAAAUCCUCAUGCACAGUGCUUUAA -((((........)))).............(((((((..((.((((((((((((.(((.((((.....)))).))).))).))))))..)))(((((.....)))))))...))))))).. ( -35.10, z-score = -3.05, R) >droPer1.super_2 8011810 120 + 9036312 -UCGUGCGCCAAAAUGAUGAAGUACAUCACGGAGCACGAGCAGGAGGAUUACCUUUUGACUGGAUUCACCAGCCAGAAGGUAAAUCCAUUCCGUGAAAAGUCUUCAUGCACCGUUCUUUAA -((((((.((....(((((.....))))).)).))))))((.((((((((((((((((.((((.....)))).)))))))).)))))..)))(((((.....)))))))............ ( -44.60, z-score = -5.06, R) >dp4.chr3 6171508 120 - 19779522 -UCGUGCGCCAAAAUGAUGAAGUAUAUCACGGAGCACGAGCAGGAGGAUUACCUUUUGACUGGAUUCACCAGCCAGAAGGUAAAUCCAUUCCGUGAAAAGUCUUCAUGCACCGUUCUUUAA -..((((..(.....)((((((....(((((((((....))....(((((((((((((.((((.....)))).)))))))).)))))..))))))).....)))))))))).......... ( -43.90, z-score = -4.97, R) >droAna3.scaffold_13266 18657203 120 - 19884421 -UCUUGCGCUAAAAUGAUGAAGUACAUCACGGAGCACGAGCAGGAGGACUACCUGCUGACUGGAUUCACCAGCCAGAAGGUUAAUCCCUUCCGCGAGAAGUCAUCAUGCACCGUACUUUAA -...((((.....(((((((..(...((.((((((....)).((.(((..((((.(((.((((.....)))).))).))))...))))))))).)).)..)))))))....))))...... ( -38.10, z-score = -2.61, R) >droEre2.scaffold_4929 8781220 120 - 26641161 -UCGUGCGCCAAGAUGAUGAAGUACAUCACGGAGCACGAGCAGGAGGACUACUUGCUCACCGGGUUCACCAGCCAGAAGGUGAAUCCGUUCCGCGAGAAGUCAUCGUGCACCGUCCUCUAA -((((((.((..((((........))))..)).))))))(((.((.((((.(((((....((((((((((........))))))))))....))))).)))).)).)))............ ( -52.30, z-score = -4.59, R) >droYak2.chr2L 17447819 120 + 22324452 -UCUUGCGCCAAGAUGAUGAAGUACAUAACGGAGCACGAGCAGGAGGACUACCUGCUCACCGGGUUCACCAGCCAGAAGGUGAAUCCCUUCCGCGAGAAGUCGUCGUGCACCGUCCUCUAA -..........(((.((((..((((.....((((...(((((((.......)))))))...(((((((((........))))))))).))))((((....)))).))))..)))).))).. ( -43.40, z-score = -2.36, R) >droSec1.super_1 2427740 120 + 14215200 -UCGUGCGCCAAGAUGAUGAAGUACAUCACGGAGCACGAGCAGGAGGAUUACCUGCUCACCGGGUUCACCAGCCAGAAGGUGAAUCCCUUCCGCGAGAAAUCGUCGUGCACCGUCCUCUAA -..((((((...((((((........((.(((((...(((((((.......)))))))...(((((((((........))))))))).))))).))...)))))))))))).......... ( -47.60, z-score = -3.61, R) >droSim1.chr2R 3463260 120 + 19596830 -UCGUGCGCCAAGAUGAUGAAGUACAUCACGGAGCACGAGCAGGAGGAUUACCUGCUUACCGGAUUCACCAGCCAGAAGGUGAAUCCCUUCCGCGAGAAAUCGUCGUGCACAGUCCUCUAA -((((((.((..((((........))))..)).))))))...((((((((...(((.....(((((((((........)))))))))....(((((....))).)).))).)))))))).. ( -48.40, z-score = -4.66, R) >apiMel3.GroupUn 246351839 120 - 399230636 GCUGU-CGAAGAUAUCAUGAAAUAUAUCACAGAACACGAACAGGAAGAUUAUCUUUUGGUUGGUUUUUCAUCACAAAAAUCAAAUCCCUUUCGUGAAAGAAGUUACUGUACCAUUUUUUAA .((((-....(((((........)))))))))..((((((.((((((.....)))....(((((((((......)))))))))...))))))))).......................... ( -19.00, z-score = 0.43, R) >triCas2.ChLG2 6125017 120 + 12900155 GCAGUG-GCCGAUAUCAUCAAGUACAUCACCGAACAUGAGCAAGACGACUGCCUCCUUGUGGGGUUCUCGAGCCAGCGGGUCAACCCCUUCAGGGAGAAGGCCCCCUGCACCCUGUUAUAG ((((.(-(((.......(((.((.(......).)).)))(((.......)))(((((((.((((((((((......))))..))))))..)))))))..))))..))))............ ( -40.70, z-score = -1.14, R) >consensus _UCGUGCGCCAAAAUGAUGAAGUACAUCACGGAGCACGAGCAGGAGGAUUACCUGUUGACUGGGUUCACCAGCCAGAAGGUAAAUCCCUUCCGCGAGAAGUCCUCGUGCACCGUUCUUUAA ..............................((((...(.((.((((...(((((.((..(.((.....)).)..)).)))))......))))((((.......)))))).)....)))).. ( -8.17 = -7.32 + -0.85)
| Location | 4,789,945 – 4,790,065 |
|---|---|
| Length | 120 |
| Sequences | 15 |
| Columns | 121 |
| Reading direction | reverse |
| Mean pairwise identity | 78.71 |
| Shannon entropy | 0.48956 |
| G+C content | 0.49389 |
| Mean single sequence MFE | -42.44 |
| Consensus MFE | -11.65 |
| Energy contribution | -11.67 |
| Covariance contribution | 0.02 |
| Combinations/Pair | 1.56 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.27 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.534743 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 4789945 120 - 21146708 UUAGAGAACGGUGCAGGACGACUUCUCGCGGAAGGGAUUCACCUUCUGGCUGGUGAACCCGGUGAGCAGGUAGUCCUCCUGCUCGUGCUCCGUGAUGUACUUCAUCAUCUUAGCGCAUGA- ...(((..(((.(.(((((.(((((((((....(((.((((((........))))))))).))))).)))).))))))))))))(((((..((((((.....))))))...)))))....- ( -48.80, z-score = -2.41, R) >anoGam1.chr3L 5195294 120 - 41284009 UUACAGAAUGCUGCAGGAGCUCUUCUCGCGGAACGGGUUCACCUUUUGGCUGGAGAACCCGACCAACAGGCAGUCUUCGGCCUGGUGCUCCGUCACGUACUUCACGAUGUCCGC-CACGGC .........((((...(((.....)))((((((((((..((((....(((((((((..((........))...))))))))).)))).)))))..(((.....)))...)))))-..)))) ( -41.20, z-score = -0.49, R) >droGri2.scaffold_15112 3867477 120 - 5172618 UUAGAGAGCCGUGCACGAGGACUUCUCACGGAAGGGGUUUACCUUUUGGCUGGUGAAACCAGUCAAUAGAUAAUCCUCCUGCUCGUGCUCCGAUAUAUAUUUCUUCAUUUUAUCGCAAGA- ...(.((((((.(((.(((((((((.....))))........((.((((((((.....)))))))).))....))))).))).)).))))).....................((....))- ( -39.80, z-score = -2.93, R) >droMoj3.scaffold_6496 13147778 120 + 26866924 UUAAAGAGCAGUGCACGACGACUUCUCACGGAAUGGGUUUACCUUCUGACUGGUGAAGCCAGUCAACAGAUAAUCCUCCUGCUCGUGCUCCGAUAUAUACUUCUUCAUUUUAUCGCACGA- .....((((((((...((.....)).)))(((..(((((...((..(((((((.....)))))))..))..)))))))))))))((((...((((...............))))))))..- ( -32.66, z-score = -1.83, R) >droVir3.scaffold_12875 1773228 120 - 20611582 UUAAAGAGCAGUGCACGAGGAUUUCUCGCGGAAGGGAUUUACCUUCUGGCUGGUGAAACCAGUCAAUAGGUAAUCCUCCUGCUCGUGCUCCGAUAUAUAUUUCUUCAUUUUGUCGCAAGA- .....((((((.(((.(((((....((.(....).)).((((((..(((((((.....)))))))..))))))))))).))).).)))))(((((...............))))).....- ( -44.16, z-score = -4.01, R) >droWil1.scaffold_180697 1470382 120 + 4168966 UUAAAGCACUGUGCAUGAGGAUUUCUCACGGAACGGAUUAACCUUUUGGCUGGUGAAACCAGUCAAUAGAUAAUCCUCCUGCUCAUGCUCUGUGAUAUAUUUCAUCAUUUUGUUGCAUGA- ......(((.(.(((((((((....))..(((..((((((..((.((((((((.....)))))))).)).)))))))))..))))))).).))).......((((((......)).))))- ( -37.40, z-score = -3.32, R) >droPer1.super_2 8011810 120 - 9036312 UUAAAGAACGGUGCAUGAAGACUUUUCACGGAAUGGAUUUACCUUCUGGCUGGUGAAUCCAGUCAAAAGGUAAUCCUCCUGCUCGUGCUCCGUGAUGUACUUCAUCAUUUUGGCGCACGA- ........((((((((((((((...(((((((..(((((.(((((.(((((((.....))))))).))))))))))....((....))))))))).)).)))))).......)))).)).- ( -47.41, z-score = -5.18, R) >dp4.chr3 6171508 120 + 19779522 UUAAAGAACGGUGCAUGAAGACUUUUCACGGAAUGGAUUUACCUUCUGGCUGGUGAAUCCAGUCAAAAGGUAAUCCUCCUGCUCGUGCUCCGUGAUAUACUUCAUCAUUUUGGCGCACGA- ........((((((((((((.....(((((((..(((((.(((((.(((((((.....))))))).))))))))))....((....)))))))))....)))))).......)))).)).- ( -45.71, z-score = -5.02, R) >droAna3.scaffold_13266 18657203 120 + 19884421 UUAAAGUACGGUGCAUGAUGACUUCUCGCGGAAGGGAUUAACCUUCUGGCUGGUGAAUCCAGUCAGCAGGUAGUCCUCCUGCUCGUGCUCCGUGAUGUACUUCAUCAUUUUAGCGCAAGA- (((((((((((.((((((.((....))((((..(((((((.(((.((((((((.....)))))))).)))))))))).)))))))))).))))((((.....)))))))))))(....).- ( -50.00, z-score = -4.64, R) >droEre2.scaffold_4929 8781220 120 + 26641161 UUAGAGGACGGUGCACGAUGACUUCUCGCGGAACGGAUUCACCUUCUGGCUGGUGAACCCGGUGAGCAAGUAGUCCUCCUGCUCGUGCUCCGUGAUGUACUUCAUCAUCUUGGCGCACGA- ...(((((((.....)....(((((((((....(((.((((((........)))))).)))))))).)))).))))))....((((((.(((.((((........)))).))).))))))- ( -47.80, z-score = -2.44, R) >droYak2.chr2L 17447819 120 - 22324452 UUAGAGGACGGUGCACGACGACUUCUCGCGGAAGGGAUUCACCUUCUGGCUGGUGAACCCGGUGAGCAGGUAGUCCUCCUGCUCGUGCUCCGUUAUGUACUUCAUCAUCUUGGCGCAAGA- ...((((((((.((((............(((((((......)))))))(((((.....)))))(((((((.......))))))))))).))))......))))....(((((...)))))- ( -45.30, z-score = -1.42, R) >droSec1.super_1 2427740 120 - 14215200 UUAGAGGACGGUGCACGACGAUUUCUCGCGGAAGGGAUUCACCUUCUGGCUGGUGAACCCGGUGAGCAGGUAAUCCUCCUGCUCGUGCUCCGUGAUGUACUUCAUCAUCUUGGCGCACGA- ...(((((((........))..))))).(((((((......)))))))(((((.....)))))(((((((.......)))))))((((.(((.((((........)))).))).))))..- ( -48.60, z-score = -2.47, R) >droSim1.chr2R 3463260 120 - 19596830 UUAGAGGACUGUGCACGACGAUUUCUCGCGGAAGGGAUUCACCUUCUGGCUGGUGAAUCCGGUAAGCAGGUAAUCCUCCUGCUCGUGCUCCGUGAUGUACUUCAUCAUCUUGGCGCACGA- ...(((((.(.(((.((.(((....)))))....(((((((((........))))))))).....))).)...)))))....((((((.(((.((((........)))).))).))))))- ( -46.60, z-score = -2.66, R) >apiMel3.GroupUn 246351839 120 + 399230636 UUAAAAAAUGGUACAGUAACUUCUUUCACGAAAGGGAUUUGAUUUUUGUGAUGAAAAACCAACCAAAAGAUAAUCUUCCUGUUCGUGUUCUGUGAUAUAUUUCAUGAUAUCUUCG-ACAGC ..........................((((((.((((.(((.((((((.(.((......)).))))))).)))...)))).))))))..((((((.(((((....)))))..)).-)))). ( -21.10, z-score = -0.01, R) >triCas2.ChLG2 6125017 120 - 12900155 CUAUAACAGGGUGCAGGGGGCCUUCUCCCUGAAGGGGUUGACCCGCUGGCUCGAGAACCCCACAAGGAGGCAGUCGUCUUGCUCAUGUUCGGUGAUGUACUUGAUGAUAUCGGC-CACUGC ......((((((.(((((((....)))))))..(((.....))))))(((.(((...((((....)).))..((((((.(((((((.....)))).)))...)))))).)))))-).))). ( -40.10, z-score = 0.65, R) >consensus UUAAAGAACGGUGCACGACGACUUCUCGCGGAAGGGAUUCACCUUCUGGCUGGUGAACCCAGUCAACAGGUAAUCCUCCUGCUCGUGCUCCGUGAUGUACUUCAUCAUUUUGGCGCACGA_ ............((((((..........(((((.((.....)))))))..(((.....))).....................))))))................................. (-11.65 = -11.67 + 0.02)
| Location | 4,789,970 – 4,790,082 |
|---|---|
| Length | 112 |
| Sequences | 15 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 72.11 |
| Shannon entropy | 0.62844 |
| G+C content | 0.52996 |
| Mean single sequence MFE | -40.14 |
| Consensus MFE | -8.17 |
| Energy contribution | -7.32 |
| Covariance contribution | -0.85 |
| Combinations/Pair | 1.72 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.20 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.845331 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
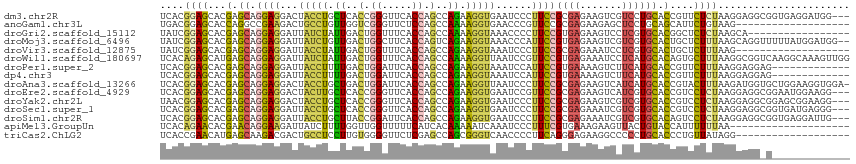
>dm3.chr2R 4789970 112 + 21146708 UCACGGAGCACGAGCAGGAGGACUACCUGCUCACCGGGUUCACCAGCCAGAAGGUGAAUCCCUUCCGCGAGAAGUCGUCCUGCACCGUUCUCUAAGGAGGCGGUGAGGAUGG--- ((.(((((...(((((((.......)))))))...(((((((((........))))))))).))))).)).....((((((.((((((.(((....))))))))))))))).--- ( -55.50, z-score = -4.22, R) >anoGam1.chr3L 5195319 96 + 41284009 UGACGGAGCACCAGGCCGAAGACUGCCUGUUGGUCGGGUUCUCCAGCCAAAAGGUGAACCCGUUCCGCGAGAAGAGCUCCUGCAGCAUUCUGUAAG------------------- ....(((((..(((((........)))))..((.(((((((.((........)).)))))))..)).........)))))(((((....)))))..------------------- ( -37.80, z-score = -2.38, R) >droGri2.scaffold_15112 3867502 98 + 5172618 UAUCGGAGCACGAGCAGGAGGAUUAUCUAUUGACUGGUUUCACCAGCCAAAAGGUAAACCCCUUCCGUGAGAAGUCCUCGUGCACGGCUCUCUAAGCA----------------- ....(((((.((.(((.(((((((((((.(((.((((.....)))).))).))))))....(((.(....))))))))).))).))))))).......----------------- ( -38.40, z-score = -3.90, R) >droMoj3.scaffold_6496 13147803 113 - 26866924 UAUCGGAGCACGAGCAGGAGGAUUAUCUGUUGACUGGCUUCACCAGUCAGAAGGUAAACCCAUUCCGUGAGAAGUCGUCGUGCACUGCUCUUUAAGCAGGUUUUUAUGGAUGG-- ((((...((((((...(((((.((((((.((((((((.....)))))))).)))))).))...)))..((....)).)))))).(((((.....))))).........)))).-- ( -38.90, z-score = -2.37, R) >droVir3.scaffold_12875 1773253 96 + 20611582 UAUCGGAGCACGAGCAGGAGGAUUACCUAUUGACUGGUUUCACCAGCCAGAAGGUAAAUCCCUUCCGCGAGAAAUCCUCGUGCACUGCUCUUUAAG------------------- ....((((((.(.(((.(((((((((((.(((.((((.....)))).))).))))))........(....)...))))).))).))))))).....------------------- ( -37.70, z-score = -4.58, R) >droWil1.scaffold_180697 1470407 115 - 4168966 UCACAGAGCAUGAGCAGGAGGAUUAUCUAUUGACUGGUUUCACCAGCCAAAAGGUUAAUCCGUUCCGUGAGAAAUCCUCAUGCACAGUGCUUUAAGGCGGUCAAGGCAAAGUUGG .......(((((((..((((((((((((.(((.((((.....)))).))).))).))))))..)))..((....)))))))))((..((((((.........))))))..))... ( -35.40, z-score = -2.07, R) >droPer1.super_2 8011835 102 + 9036312 UCACGGAGCACGAGCAGGAGGAUUACCUUUUGACUGGAUUCACCAGCCAGAAGGUAAAUCCAUUCCGUGAAAAGUCUUCAUGCACCGUUCUUUAAGGAGGAG------------- (((((((((....))....(((((((((((((.((((.....)))).)))))))).)))))..)))))))....(((((.................))))).------------- ( -36.73, z-score = -3.84, R) >dp4.chr3 6171533 102 - 19779522 UCACGGAGCACGAGCAGGAGGAUUACCUUUUGACUGGAUUCACCAGCCAGAAGGUAAAUCCAUUCCGUGAAAAGUCUUCAUGCACCGUUCUUUAAGGAGGAG------------- (((((((((....))....(((((((((((((.((((.....)))).)))))))).)))))..)))))))....(((((.................))))).------------- ( -36.73, z-score = -3.84, R) >droAna3.scaffold_13266 18657228 114 - 19884421 UCACGGAGCACGAGCAGGAGGACUACCUGCUGACUGGAUUCACCAGCCAGAAGGUUAAUCCCUUCCGCGAGAAGUCAUCAUGCACCGUACUUUAAGAUGGUGCUGGAAGGUGGA- ((.((((((....)).((.(((..((((.(((.((((.....)))).))).))))...))))))))).))....(((((..(((((((........))))))).....))))).- ( -38.60, z-score = -1.26, R) >droEre2.scaffold_4929 8781245 112 - 26641161 UCACGGAGCACGAGCAGGAGGACUACUUGCUCACCGGGUUCACCAGCCAGAAGGUGAAUCCGUUCCGCGAGAAGUCAUCGUGCACCGUCCUCUAAGGAGGCGGAAUGGAAGG--- ((.((((((..(((((((.......)))))))...(((((((((........))))))))))))))).)).......((((...(((.((((....))))))).))))....--- ( -48.50, z-score = -3.36, R) >droYak2.chr2L 17447844 112 + 22324452 UAACGGAGCACGAGCAGGAGGACUACCUGCUCACCGGGUUCACCAGCCAGAAGGUGAAUCCCUUCCGCGAGAAGUCGUCGUGCACCGUCCUCUAAGGAGGCGGAGCGGAAGG--- ...........(((((((.......)))))))...(((((((((........)))))))))((((((((((....).))))((.(((.((((....))))))).))))))).--- ( -51.00, z-score = -3.18, R) >droSec1.super_1 2427765 112 + 14215200 UCACGGAGCACGAGCAGGAGGAUUACCUGCUCACCGGGUUCACCAGCCAGAAGGUGAAUCCCUUCCGCGAGAAAUCGUCGUGCACCGUCCUCUAAGGAGGCGGUGAUGAGGG--- ((.(((((...(((((((.......)))))))...(((((((((........))))))))).))))).)).....(.((((.(((((.((((....))))))))))))).).--- ( -53.80, z-score = -4.33, R) >droSim1.chr2R 3463285 112 + 19596830 UCACGGAGCACGAGCAGGAGGAUUACCUGCUUACCGGAUUCACCAGCCAGAAGGUGAAUCCCUUCCGCGAGAAAUCGUCGUGCACAGUCCUCUAAGGAGGCGGUGAGGAUUG--- .......(((((((((((.......)))))))...(((((((((........))))))))).....((((....)))).)))).((((((((...(....)...))))))))--- ( -46.40, z-score = -3.46, R) >apiMel3.GroupUn 246351864 95 - 399230636 UCACAGAACACGAACAGGAAGAUUAUCUUUUGGUUGGUUUUUCAUCACAAAAAUCAAAUCCCUUUCGUGAAAGAAGUUACUGUACCAUUUUUUAA-------------------- ..((((..((((((.((((((.....)))....(((((((((......)))))))))...)))))))))((.....)).))))............-------------------- ( -15.20, z-score = 0.02, R) >triCas2.ChLG2 6125042 96 + 12900155 UCACCGAACAUGAGCAAGACGACUGCCUCCUUGUGGGGUUCUCGAGCCAGCGGGUCAACCCCUUCAGGGAGAAGGCCCCCUGCACCCUGUUAUAGG------------------- ...((.((((.(.(((.(....((..(((((((.((((((((((......))))..))))))..))))))).))....).)))..).))))...))------------------- ( -31.40, z-score = 0.13, R) >consensus UCACGGAGCACGAGCAGGAGGAUUACCUGUUGACUGGGUUCACCAGCCAGAAGGUAAAUCCCUUCCGCGAGAAGUCCUCGUGCACCGUUCUUUAAGGAGG_G_____________ ....((((...(.((.((((...(((((.((..(.((.....)).)..)).)))))......))))((((.......)))))).)....))))...................... ( -8.17 = -7.32 + -0.85)
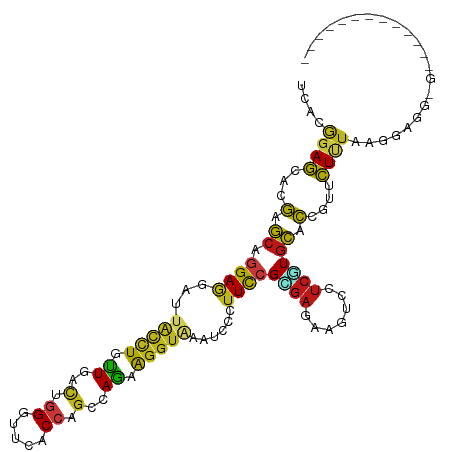
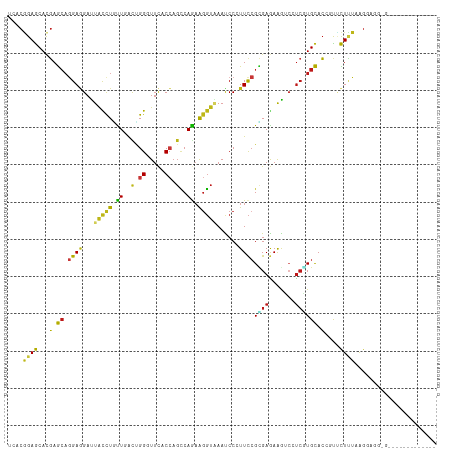
| Location | 4,789,970 – 4,790,082 |
|---|---|
| Length | 112 |
| Sequences | 15 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 72.11 |
| Shannon entropy | 0.62844 |
| G+C content | 0.52996 |
| Mean single sequence MFE | -39.59 |
| Consensus MFE | -11.65 |
| Energy contribution | -11.67 |
| Covariance contribution | 0.02 |
| Combinations/Pair | 1.56 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.29 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.973627 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
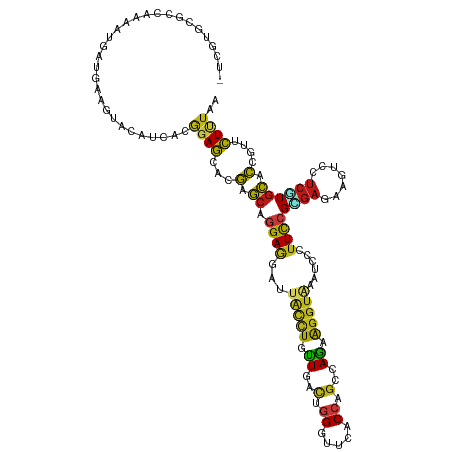
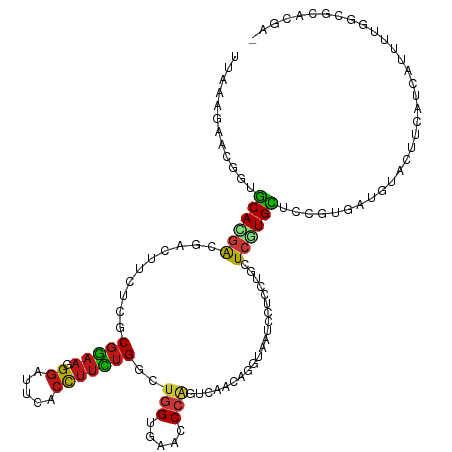
>dm3.chr2R 4789970 112 - 21146708 ---CCAUCCUCACCGCCUCCUUAGAGAACGGUGCAGGACGACUUCUCGCGGAAGGGAUUCACCUUCUGGCUGGUGAACCCGGUGAGCAGGUAGUCCUCCUGCUCGUGCUCCGUGA ---...(((((((((.(((....)))..))))).)))).......(((((((.(((.((((((........)))))))))...(((((((.......)))))))....))))))) ( -51.00, z-score = -3.11, R) >anoGam1.chr3L 5195319 96 - 41284009 -------------------CUUACAGAAUGCUGCAGGAGCUCUUCUCGCGGAACGGGUUCACCUUUUGGCUGGAGAACCCGACCAACAGGCAGUCUUCGGCCUGGUGCUCCGUCA -------------------....(((....)))..(((((.(.......((..(((((((.((........)).))))))).))..(((((........)))))).))))).... ( -36.00, z-score = -1.24, R) >droGri2.scaffold_15112 3867502 98 - 5172618 -----------------UGCUUAGAGAGCCGUGCACGAGGACUUCUCACGGAAGGGGUUUACCUUUUGGCUGGUGAAACCAGUCAAUAGAUAAUCCUCCUGCUCGUGCUCCGAUA -----------------......(.((((((.(((.(((((((((.....))))........((.((((((((.....)))))))).))....))))).))).)).))))).... ( -38.00, z-score = -3.10, R) >droMoj3.scaffold_6496 13147803 113 + 26866924 --CCAUCCAUAAAAACCUGCUUAAAGAGCAGUGCACGACGACUUCUCACGGAAUGGGUUUACCUUCUGACUGGUGAAGCCAGUCAACAGAUAAUCCUCCUGCUCGUGCUCCGAUA --..(((.........((((((...)))))).(((((((((....))..(((..(((((...((..(((((((.....)))))))..))..)))))))).).))))))...))). ( -33.40, z-score = -2.66, R) >droVir3.scaffold_12875 1773253 96 - 20611582 -------------------CUUAAAGAGCAGUGCACGAGGAUUUCUCGCGGAAGGGAUUUACCUUCUGGCUGGUGAAACCAGUCAAUAGGUAAUCCUCCUGCUCGUGCUCCGAUA -------------------......((((((.(((.(((((....((.(....).)).((((((..(((((((.....)))))))..))))))))))).))).).)))))..... ( -42.10, z-score = -4.81, R) >droWil1.scaffold_180697 1470407 115 + 4168966 CCAACUUUGCCUUGACCGCCUUAAAGCACUGUGCAUGAGGAUUUCUCACGGAACGGAUUAACCUUUUGGCUGGUGAAACCAGUCAAUAGAUAAUCCUCCUGCUCAUGCUCUGUGA ..........................(((.(.(((((((((....))..(((..((((((..((.((((((((.....)))))))).)).)))))))))..))))))).).))). ( -35.70, z-score = -3.28, R) >droPer1.super_2 8011835 102 - 9036312 -------------CUCCUCCUUAAAGAACGGUGCAUGAAGACUUUUCACGGAAUGGAUUUACCUUCUGGCUGGUGAAUCCAGUCAAAAGGUAAUCCUCCUGCUCGUGCUCCGUGA -------------..............((((.((((((.((....))..(((..(((((.(((((.(((((((.....))))))).)))))))))))))...)))))).)))).. ( -39.50, z-score = -5.00, R) >dp4.chr3 6171533 102 + 19779522 -------------CUCCUCCUUAAAGAACGGUGCAUGAAGACUUUUCACGGAAUGGAUUUACCUUCUGGCUGGUGAAUCCAGUCAAAAGGUAAUCCUCCUGCUCGUGCUCCGUGA -------------..............((((.((((((.((....))..(((..(((((.(((((.(((((((.....))))))).)))))))))))))...)))))).)))).. ( -39.50, z-score = -5.00, R) >droAna3.scaffold_13266 18657228 114 + 19884421 -UCCACCUUCCAGCACCAUCUUAAAGUACGGUGCAUGAUGACUUCUCGCGGAAGGGAUUAACCUUCUGGCUGGUGAAUCCAGUCAGCAGGUAGUCCUCCUGCUCGUGCUCCGUGA -.........................(((((.((((((.((....))((((..(((((((.(((.((((((((.....)))))))).)))))))))).)))))))))).))))). ( -45.40, z-score = -3.61, R) >droEre2.scaffold_4929 8781245 112 + 26641161 ---CCUUCCAUUCCGCCUCCUUAGAGGACGGUGCACGAUGACUUCUCGCGGAACGGAUUCACCUUCUGGCUGGUGAACCCGGUGAGCAAGUAGUCCUCCUGCUCGUGCUCCGUGA ---............((((....))))((((.((((((.((((.((.((....(((.((((((........)))))).)))....)).)).)))).......)))))).)))).. ( -41.60, z-score = -1.81, R) >droYak2.chr2L 17447844 112 - 22324452 ---CCUUCCGCUCCGCCUCCUUAGAGGACGGUGCACGACGACUUCUCGCGGAAGGGAUUCACCUUCUGGCUGGUGAACCCGGUGAGCAGGUAGUCCUCCUGCUCGUGCUCCGUUA ---((((((((.(((((((....)))).))).))..(.(((....))))))))))..((((((........))))))..(((((((((((.......))))))))....)))... ( -48.40, z-score = -2.38, R) >droSec1.super_1 2427765 112 - 14215200 ---CCCUCAUCACCGCCUCCUUAGAGGACGGUGCACGACGAUUUCUCGCGGAAGGGAUUCACCUUCUGGCUGGUGAACCCGGUGAGCAGGUAAUCCUCCUGCUCGUGCUCCGUGA ---.......(((((((((....)))).)))))............(((((((.(((.((((((........)))))))))...(((((((.......)))))))....))))))) ( -50.00, z-score = -3.48, R) >droSim1.chr2R 3463285 112 - 19596830 ---CAAUCCUCACCGCCUCCUUAGAGGACUGUGCACGACGAUUUCUCGCGGAAGGGAUUCACCUUCUGGCUGGUGAAUCCGGUAAGCAGGUAAUCCUCCUGCUCGUGCUCCGUGA ---.......(((.(((((....)))).).)))............(((((((..(((((((((........))))))))).(((((((((.......))))))..)))))))))) ( -42.30, z-score = -2.38, R) >apiMel3.GroupUn 246351864 95 + 399230636 --------------------UUAAAAAAUGGUACAGUAACUUCUUUCACGAAAGGGAUUUGAUUUUUGUGAUGAAAAACCAACCAAAAGAUAAUCUUCCUGUUCGUGUUCUGUGA --------------------...........(((((..........((((((.((((.(((.((((((.(.((......)).))))))).)))...)))).))))))..))))). ( -19.20, z-score = -1.08, R) >triCas2.ChLG2 6125042 96 - 12900155 -------------------CCUAUAACAGGGUGCAGGGGGCCUUCUCCCUGAAGGGGUUGACCCGCUGGCUCGAGAACCCCACAAGGAGGCAGUCGUCUUGCUCAUGUUCGGUGA -------------------((...((((..(.(((((((((..(((((.((..((((((((((....)).))...)))))).)).)))))..))).)))))).).)))).))... ( -31.80, z-score = 1.35, R) >consensus _____________C_CCUCCUUAAAGAACGGUGCACGACGACUUCUCGCGGAAGGGAUUCACCUUCUGGCUGGUGAACCCAGUCAACAGGUAAUCCUCCUGCUCGUGCUCCGUGA ................................((((((..........(((((.((.....)))))))..(((.....))).....................))))))....... (-11.65 = -11.67 + 0.02)
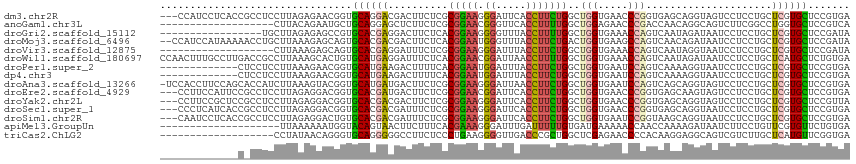
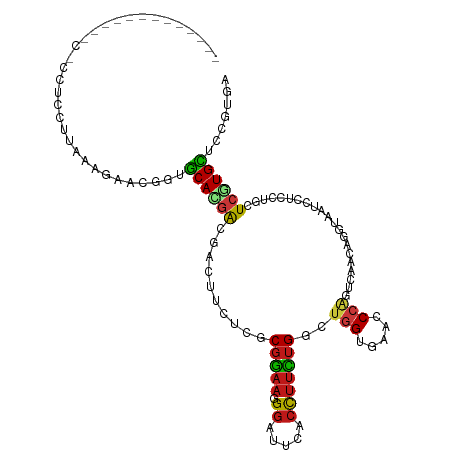
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:09:19 2011