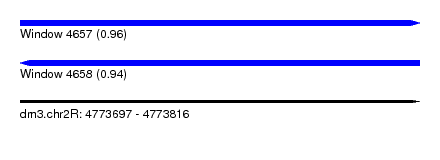
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 4,773,697 – 4,773,816 |
| Length | 119 |
| Max. P | 0.964267 |

| Location | 4,773,697 – 4,773,816 |
|---|---|
| Length | 119 |
| Sequences | 7 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 80.31 |
| Shannon entropy | 0.40037 |
| G+C content | 0.59755 |
| Mean single sequence MFE | -49.33 |
| Consensus MFE | -29.49 |
| Energy contribution | -31.86 |
| Covariance contribution | 2.37 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.964267 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 4773697 119 + 21146708 AAGAAGGCGGCACAGGAGCGGCAGGAGGCGCAAAAGGCUCAGGAGGCCGCUAUGGCCUCCAUCGAGGCGGAAGCCAAGAAGUCGUCAUCUGCGCCUCAGGAGACUCCAGCUGUUCACCA ........((.....(((((((..((((((((...(((...(((((((.....))))))).((..(((....)))..))....)))...))))))))((....))...))))))).)). ( -57.10, z-score = -3.35, R) >droSim1.chr2R 3446648 119 + 19596830 AAGAAGGCGGCACAGGAGCGGCAGGAGGCGCAAAAGGCUCAGGAGGCCGCUAUGGCCUCCAUCGAGGCGGAAGCCAAGAAGUCGUCAUCUGCGCCUCAGGAGACUCCAGCCGUCCACCA .....((((((...(((((..(..((((((((...(((...(((((((.....))))))).((..(((....)))..))....)))...))))))))..).).)))).))))))..... ( -60.60, z-score = -4.15, R) >droSec1.super_1 2411549 119 + 14215200 AAGAAGGCGGCACAAGAGCGGCAGGAGGCGCAAAAGGCUCAGGAGGCCGCUAUGGCCUCCAUCGAGGCGGAAGCCAAGAAGUCGUCAUCUGCGCCUCAGGAGACUCCAGCCGUCCACCA .....((.((.......(((((..((((((((...(((...(((((((.....))))))).((..(((....)))..))....)))...))))))))((....))...))))))).)). ( -57.71, z-score = -3.70, R) >droYak2.chr2L 17432178 119 + 22324452 AAGAAGGCGGCACAGGAGCGGCAGGAAGCACAAAAGGCUCAGGAGGCCGCGAUGGCCUCCAUCGAGGCGGAAGCCAAGAAGUCGUCAUCUGCGCCUCAGGAGACUCCAGCCGUCCACCA ..((.((((((.((((.(((((..((.((.......)))).(((((((.....))))))).((..(((....)))..)).)))))..)))).)))..((....))...))).))..... ( -50.10, z-score = -2.02, R) >droEre2.scaffold_4929 8763395 119 - 26641161 AAGAAGGCGGCACAGGAGCGGCAGGAGGCGCAAAAGGCUCAGGAGGCCGCAAUGGCCUCCAUCGAGGCGGAAGCCAAGAAGUCCUCAUCUGCGCCUCAGGAGACUCCAGCCGUCCACCA .....((((((...(((((..(..((((((((..(((((..(((((((.....))))))).((..(((....)))..)))).)))....))))))))..).).)))).))))))..... ( -60.40, z-score = -4.05, R) >droWil1.scaffold_180697 1453561 119 - 4168966 AAAAAGGCUGCCCAAGAGCGUCAGGAGGCUCAGAAGGCACAAGAAGCGGCUAUGGCAUCGAUUGAAGCGGAGGCCAAGAAAUCCUCUGCUGCCCCCCAGGAGACUCCAGCUGUUCAUCA ........((((...((((.(....).))))....))))...(((.(((((..(((.((....))((((((((.........)))))))))))....((....))..)))))))).... ( -35.50, z-score = 1.17, R) >triCas2.ChLG3 11743028 113 - 32080666 AAACGAGCAGCUCAAGAAAAAGUCGAAGCUCAAGCGGCUCAACAAGCGGUAAUGCAAGCAAUUGAGUCCGAAUCCAAACA------ACCUGUUGUUACUGAACCUUCUGCCGUUUCACA (((((.((((...(((......(((..((((((((.(((.....))).))..((....)).)))))).))).....((((------(....))))).......)))))))))))).... ( -23.90, z-score = 0.24, R) >consensus AAGAAGGCGGCACAGGAGCGGCAGGAGGCGCAAAAGGCUCAGGAGGCCGCUAUGGCCUCCAUCGAGGCGGAAGCCAAGAAGUCGUCAUCUGCGCCUCAGGAGACUCCAGCCGUCCACCA .....((((((....(((..(((((((.(......).))).(((((((.....))))))).((..(((....)))..)).........))))..))).((.....)).))))))..... (-29.49 = -31.86 + 2.37)

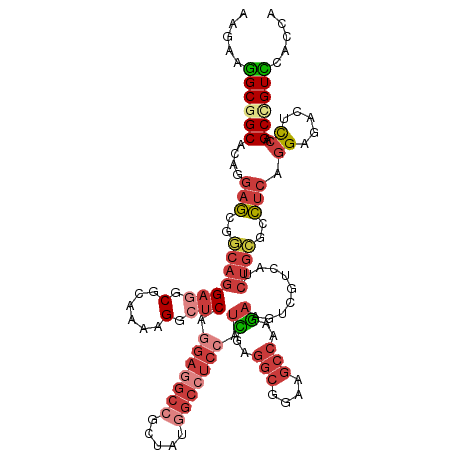
| Location | 4,773,697 – 4,773,816 |
|---|---|
| Length | 119 |
| Sequences | 7 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 80.31 |
| Shannon entropy | 0.40037 |
| G+C content | 0.59755 |
| Mean single sequence MFE | -48.36 |
| Consensus MFE | -36.05 |
| Energy contribution | -36.29 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.942714 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 4773697 119 - 21146708 UGGUGAACAGCUGGAGUCUCCUGAGGCGCAGAUGACGACUUCUUGGCUUCCGCCUCGAUGGAGGCCAUAGCGGCCUCCUGAGCCUUUUGCGCCUCCUGCCGCUCCUGUGCCGCCUUCUU .((((....((.(((((.....((((((((((.(.(....((..(((....)))..)).(((((((.....)))))))...).).)))))))))).....))))).))..))))..... ( -51.40, z-score = -2.53, R) >droSim1.chr2R 3446648 119 - 19596830 UGGUGGACGGCUGGAGUCUCCUGAGGCGCAGAUGACGACUUCUUGGCUUCCGCCUCGAUGGAGGCCAUAGCGGCCUCCUGAGCCUUUUGCGCCUCCUGCCGCUCCUGUGCCGCCUUCUU .((.((.((((.(((((.....((((((((((.(.(....((..(((....)))..)).(((((((.....)))))))...).).)))))))))).....)))))...)))))).)).. ( -55.80, z-score = -2.99, R) >droSec1.super_1 2411549 119 - 14215200 UGGUGGACGGCUGGAGUCUCCUGAGGCGCAGAUGACGACUUCUUGGCUUCCGCCUCGAUGGAGGCCAUAGCGGCCUCCUGAGCCUUUUGCGCCUCCUGCCGCUCUUGUGCCGCCUUCUU .((.((.((((.(((((.....((((((((((.(.(....((..(((....)))..)).(((((((.....)))))))...).).)))))))))).....)))))...)))))).)).. ( -53.90, z-score = -2.52, R) >droYak2.chr2L 17432178 119 - 22324452 UGGUGGACGGCUGGAGUCUCCUGAGGCGCAGAUGACGACUUCUUGGCUUCCGCCUCGAUGGAGGCCAUCGCGGCCUCCUGAGCCUUUUGUGCUUCCUGCCGCUCCUGUGCCGCCUUCUU .((.((.((((.(((((.....((((((((((.(.(....((..(((....)))..)).(((((((.....)))))))...).).)))))))))).....)))))...)))))).)).. ( -51.20, z-score = -2.04, R) >droEre2.scaffold_4929 8763395 119 + 26641161 UGGUGGACGGCUGGAGUCUCCUGAGGCGCAGAUGAGGACUUCUUGGCUUCCGCCUCGAUGGAGGCCAUUGCGGCCUCCUGAGCCUUUUGCGCCUCCUGCCGCUCCUGUGCCGCCUUCUU .((.((.((((.(((((.....((((((((((..(((.(((...(((....))).....(((((((.....))))))).)))))))))))))))).....)))))...)))))).)).. ( -58.80, z-score = -3.37, R) >droWil1.scaffold_180697 1453561 119 + 4168966 UGAUGAACAGCUGGAGUCUCCUGGGGGGCAGCAGAGGAUUUCUUGGCCUCCGCUUCAAUCGAUGCCAUAGCCGCUUCUUGUGCCUUCUGAGCCUCCUGACGCUCUUGGGCAGCCUUUUU .........(((((((.(((..((((((((..(((((......((((.((..........)).))))......)))))..))))))))))).))))....(((....))))))...... ( -38.20, z-score = 1.30, R) >triCas2.ChLG3 11743028 113 + 32080666 UGUGAAACGGCAGAAGGUUCAGUAACAACAGGU------UGUUUGGAUUCGGACUCAAUUGCUUGCAUUACCGCUUGUUGAGCCGCUUGAGCUUCGACUUUUUCUUGAGCUGCUCGUUU ....(((((((((.((((((((.(((((....)------))))......(((.((((((.....((......))..))))))))).))))))))...(((......))))))).))))) ( -29.20, z-score = 0.47, R) >consensus UGGUGGACGGCUGGAGUCUCCUGAGGCGCAGAUGACGACUUCUUGGCUUCCGCCUCGAUGGAGGCCAUAGCGGCCUCCUGAGCCUUUUGCGCCUCCUGCCGCUCCUGUGCCGCCUUCUU .......((((.(((((.....((((((((((...........(((((((((......)))))))))....((((....).))).)))))))))).....)))))...))))....... (-36.05 = -36.29 + 0.23)
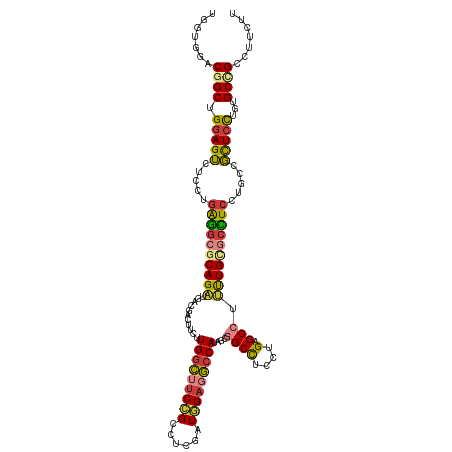
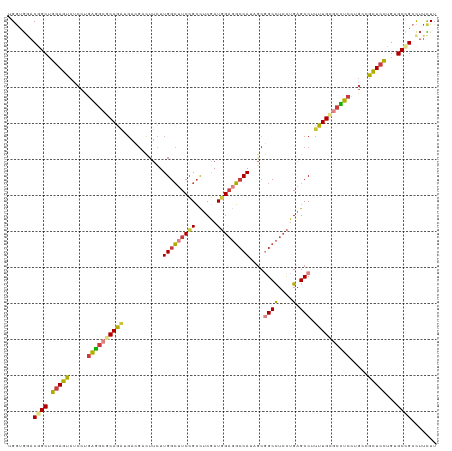
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:09:16 2011