| Sequence ID | dm3.chr2R |
|---|---|
| Location | 4,767,551 – 4,767,671 |
| Length | 120 |
| Max. P | 0.896003 |

| Location | 4,767,551 – 4,767,671 |
|---|---|
| Length | 120 |
| Sequences | 15 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.13 |
| Shannon entropy | 0.36997 |
| G+C content | 0.49000 |
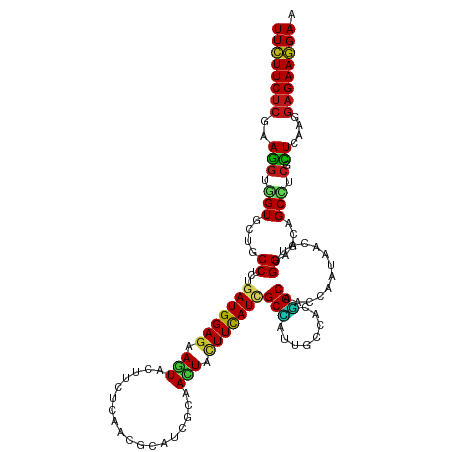
| Mean single sequence MFE | -38.65 |
| Consensus MFE | -21.10 |
| Energy contribution | -20.48 |
| Covariance contribution | -0.62 |
| Combinations/Pair | 1.45 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.896003 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
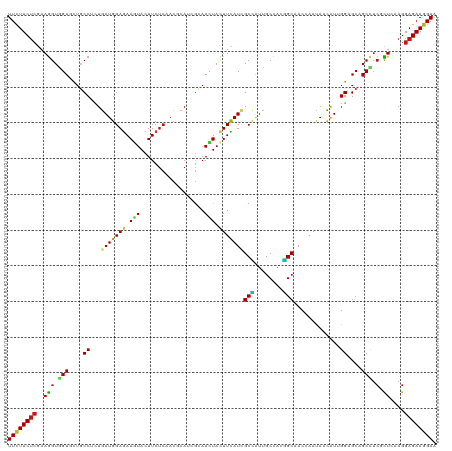
>dm3.chr2R 4767551 120 + 21146708 UUCUUCUCGAAGGUGGUGCUGCCUCUAAUGGAGAAGUACUUCUCAACGCAUCGCAACUACUUCAUCGCCAUUGCCACGGCAACCAAUAAUAUUGGAGCAGCCUCGCUCAAGGAGAAGGAA ((((((((...(((((.(((((.((((((((((((....)))))......................(((........))).........)))))))))))).)))))....)))))))). ( -39.50, z-score = -2.52, R) >droSim1.chr2R 3440526 120 + 19596830 UUCUUCUCGAAGGUGGUGCUGCCUCUGAUGGAGAAGUACUUCUCAACGCAUCGCAACUACUUCAUCGCCAUUGCCACGGCAACCAAUAAUAUUGGAGCGGCCUCGCUCAAGGAGAAGGAA ((((((((...(((((.(((((....(((((((((....)))))...((...))........))))(((........)))..(((((...))))).))))).)))))....)))))))). ( -38.60, z-score = -1.66, R) >droSec1.super_1 2405419 120 + 14215200 UUCUUCUCGAAGGUGGUGCUGCCUCUGAUGGAGAAGUACUUCUCAACGCAUCGCAACUACUUCAUCGCCAUUGCCACGGCAACCAAUAAUAUUGGCGCAGCCUCGCUCAAGGAGAAGGAA ((((((((...(((((.(((((....(((((((((....)))))...((...))........))))((((((((....))))..........))))))))).)))))....)))))))). ( -41.00, z-score = -2.51, R) >droYak2.chr2L 17426064 120 + 22324452 UUCUUCUCGAAGGUGGUGCUGCCCCUGAUGGAGAAGUACUUCUCAACGCACCGGAACUACUUUAUCGCCAUUGCCACGGCAACCAAUAAUAUUGGAGCAGCCUCGCUCAAGGAGAAGGAA ((((((((...(((((.(((((.((.(((((.((((((.((((.........)))).))))))....)))))((....)).............)).))))).)))))....)))))))). ( -39.50, z-score = -1.99, R) >droEre2.scaffold_4929 8757244 120 - 26641161 UUCUUCUCGAAGGUGGUGCUGCCCCUGAUGGAGAAGUACUUCUCAACGCACCGCAACUACUUCAUCGCCAUUGCCACGGCAACCAAUAAUAUUGGAGCAGCCUCACUCAAGGAGAAGGAA ((((((((...(((((.(((((.((.(((((((((....)))))...((...))........))))(((........))).............)).))))).)))))....)))))))). ( -40.50, z-score = -2.73, R) >droAna3.scaffold_13266 18631871 120 - 19884421 UUCUUCUCGAAGGUGGUGCUGCCCCUGAUGGAGAAAUACUUCUCCACCCACAGGAACUACUUCAUCGCCAUUGCCACGGCGACGAACAACAUUGGAGCUGCCUCCCUUAAGGAGAAGGAG ((((((((.((((.((((((.((((((.(((((((....)))))))....))))......(((.(((((........))))).))).......))))).)))..))))...)))))))). ( -45.00, z-score = -3.79, R) >dp4.chr3 13915220 120 + 19779522 UUCUUCUCGAAGGUGGUCCUGCCACUGAUGGAGAAGUACUUCUCGACGCACCGCAACUACUUCAUUGCCAUUGCCACGGCCACGAACAACAUUGGAGCUGCCUCGCUCAAGGAGAAGGAA ((((((((....((((((.((.((.((...(((((....)))))........((((........)))))).)).)).))))))...........((((......))))...)))))))). ( -36.00, z-score = -0.83, R) >droPer1.super_4 5759073 120 - 7162766 UUCUUCUCGAAGGUGGUCCUGCCACUGAUGGAGAAGUACUUCUCGACGCACCGCAACUACUUCAUUGCCAUUGCCACGGCCACGAACAACAUUGGAGCGGCCUCGCUCAAGGAGAAGGAA ((((((((....((((((.((.((.((...(((((....)))))........((((........)))))).)).)).))))))...........(((((....)))))...)))))))). ( -38.00, z-score = -1.14, R) >droWil1.scaffold_180697 1446301 120 - 4168966 UUCUUCUCGAAGGUCGUUCUGCCGCUGAUGGAGAAAUAUUUCUCAACGCAUCGUAAUUAUUUCAUAGCCAUUGCCACGGCCACUAACAACAUCGGAGCAGCCUCACUAAAGGAGAAGGAA ((((((((...(((....((((..(((((((((((....)))))......................(((........))).........)))))).))))....)))....)))))))). ( -31.00, z-score = -1.17, R) >droVir3.scaffold_12875 1751121 120 + 20611582 UUUUUCUCAAAGGUGGUGCUGCCUCUGAUGGAAAAGUACUUCUCAACCCAUCGCAAUUACUUUAUUGCCAUUGCCACUGCCACAAACAACAUAGGCGCCGCUUCGCUGAAGGAGAAAGAA ((((((((...((..(((..((....(((((...((.....))....)))))(((((......)))))....)))))..))............((((......))))....)))))))). ( -31.40, z-score = -1.14, R) >droMoj3.scaffold_6496 13125887 120 - 26866924 UUCUUCUCGAAGGUCGUGUUGCCUCUGAUGGAGAAAUAUUUCUCGACGCAUCGCAAUUAUUUUAUCGCUAUUGCCACGGCAACGAAUAACAUAGGCGCCGCAUCGCUAAAGGAGAAGGAA ((((((((....((..(((((((...(((((((((....)))))....))))(((((............)))))...)))))))....))...((((......))))....)))))))). ( -33.30, z-score = -0.75, R) >droGri2.scaffold_15112 3842908 120 + 5172618 UUCUUCUCAAAAGUGGUGCUGCCACUGAUGGAGAAAUACUUCUCGACGCAUCGCAAUUAUUUUAUUGCCAUUGCAACGGCCACAAAUAAUGUGGGUGCUGCCUCCUUGAAGGAGAAGGAA ((((((((...(((((.....)))))(((((((((....)))))........(((((......)))))))))(((.((.(((((.....))))).)).)))..........)))))))). ( -38.80, z-score = -2.57, R) >anoGam1.chr3L 8430651 120 + 41284009 UUCUUCUCCAAGGUGGUACUGCCGCUUAUGGAGAAAUAUUUCUCCAGCAAUCGUAAUUACUUCAUUGCGAUCGCUACCGCCACCAACAACGUUGGUGCGGCUUCACUGAAGGAGAAGGAA (((((((((..(((((....(((((....((((((....))))))(((.((((((((......)))))))).)))......((((((...))))))))))).)))))...))))))))). ( -52.40, z-score = -6.75, R) >apiMel3.Group2 3463384 120 + 14465785 UUCUUCAGCAAGGUCGUUCUUCCGUUGAUGGAAAAAUAUUUCAGCACGCACAAGAAUUACUUCAUCGCUGUGGCCACGGCUACCAACAACGUUGGAGCUGCGUCUCUGAAGGAGAAAGAG ((((((((..(.((.(((((..(((((.(((..........((((......(((.....)))....))))((((....))))))).)))))..))))).)).)..))))))))....... ( -36.70, z-score = -1.86, R) >triCas2.ChLG3 11719701 120 - 32080666 UUCUUCUCGAAGGUCGUCCUCCCUCUGAUGGAGAAAUAUUUCUCCACUCAUCGCAACUACUUCAUUGCUAUUGCUACAGCUACGAAUAAUGUCGGAGCUGCUAGUUUGAAGGAGAAGGAA (((((((((((((....))).....((((((((((....))))))).))))).......(((((........(((((((((.(((......))).))))).)))).))))))))))))). ( -38.10, z-score = -2.75, R) >consensus UUCUUCUCGAAGGUGGUGCUGCCUCUGAUGGAGAAGUACUUCUCAACGCAUCGCAACUACUUCAUCGCCAUUGCCACGGCAACCAAUAACAUUGGAGCAGCCUCGCUCAAGGAGAAGGAA ((((((((..(((.(((....((...(((((((.(((..................))).)))))))(((........))).............))....))).).))....)))))))). (-21.10 = -20.48 + -0.62)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:09:10 2011