
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 4,755,937 – 4,756,071 |
| Length | 134 |
| Max. P | 0.716387 |

| Location | 4,755,937 – 4,756,057 |
|---|---|
| Length | 120 |
| Sequences | 15 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.89 |
| Shannon entropy | 0.47462 |
| G+C content | 0.51661 |
| Mean single sequence MFE | -39.51 |
| Consensus MFE | -18.72 |
| Energy contribution | -18.42 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.79 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.716387 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 4755937 120 + 21146708 CGUGCUCAAUCUGAUCCUGGAGGCCAUUGACAAGAUCAACAUAAUCACCUCGCAGGGCUUCCUAGCCAGCUUCCUGGCCGGCGACGAGACCGGCCAGAGCUGGGAUCUCAUCUCCACCUA .(((........(((((.(((((((..(((...(((.......)))...)))...)))))))....(((((..(((((((((.....).)))))))))))))))))).......)))... ( -45.86, z-score = -2.40, R) >droSim1.chr2R 3433080 120 + 19596830 CGUGCUCAAUCUGAUCCUGGAGGCCAUUGACAAGAUCAACAUAAUCACCUCGCAGGGCUUCCUCGCCAGCGUGGUGGCCGGCGACGAGACCGGCCAGAGCUGGGAUCUCAUUUCCACCUA .(((...(((..(((((.(((((((..(((...(((.......)))...)))...)))))))....((((....((((((((.....).)))))))..)))))))))..)))..)))... ( -42.90, z-score = -0.95, R) >droSec1.super_1 2393735 120 + 14215200 CGUGCUCAAUCUGAUCCUAGAGGCCAUUGACAAGAUUAACAUAAUCACCUCGCAGGGCUUCCUCGCCAGCUUCCUGGCCGGCGACGAGACCGGCCAGAGCUGGGAUCUCAUUUCCACCUA .(((...(((..(((((..((((((..(((...(((((...)))))...)))...)))))).....(((((..(((((((((.....).))))))))))))))))))..)))..)))... ( -44.60, z-score = -2.86, R) >droYak2.chr2L 17414498 120 + 22324452 CGUGCUCAAUCUGAUCCUGGAGGCCAUUGACAAGAUCAACAUUAUCACCUCGCAGGGCUUUCUCGCCAGCUUUCUGGCUGGCGACGAAACCGGCCAGAGCUGGGAUCUCAUCUCCACCUA .(((........(((((..(.((((.((((.....))))................((.(((((((((((((....))))))))).))))))))))....)..))))).......)))... ( -43.76, z-score = -2.54, R) >droEre2.scaffold_4929 8745632 120 - 26641161 CGUGCUGAAUCUGAUCCUGGAGGCCAUUGACAAGAUCAACAUAAUCACCUCGCAGGGCUUCCUCGCCAGCUUCCUGGCCGGCGACGAGACCGGUCAGAGCUGGGAUCUCAUCUCCACCUA .(((..((....(((((.(((((((..(((...(((.......)))...)))...)))))))....(((((..(((((((((.....).))))))))))))))))))...))..)))... ( -45.30, z-score = -1.96, R) >droAna3.scaffold_13266 18620866 120 - 19884421 AGUUCUCAACCUCAUCCUGGAGGCUAUUGACAAGAUCAAUAUAAUUACAUCUCAGGGCUUCCUGGCCAGCUUCCUGGCCGGAGACGAGACUGGCCAAAGCUGGGACCUCAUCUCCACCUA ((((((...((((......)))).((((((.....))))))............))))))....((((((....))))))(((((.(((.(..((....))..)...))).)))))..... ( -38.80, z-score = -1.39, R) >dp4.chr3 13904179 120 + 19779522 CGUUCUCAAUCUUAUACUGGAGGCCAUUGACAAGAUCAAUAUUAUCACGUCGCAGGGCUUCCUGGCCAGCUUUCUCGCUGGCGACGAGACCGGCCAAAGCUGGGACCUCAUCUCCACGUA (((((((.........(.(((((((..((((..(((.......)))..))))...))))))).)((((((......))))))...))))(((((....)))))............))).. ( -41.00, z-score = -2.39, R) >droPer1.super_4 5747885 120 - 7162766 CGUUCUCAAUCUUAUACUGGAGGCCAUUGACAAGAUCAAUAUCAUCACGUCGCAGGGCUUCCUGGCCAGCUUUCUCGCUGGCGACGAGACCGGCCAAAGCUGGGACCUCAUCUCCACGUA (((((((.........(.(((((((..((((..(((.......)))..))))...))))))).)((((((......))))))...))))(((((....)))))............))).. ( -41.10, z-score = -2.30, R) >droWil1.scaffold_180697 1435918 120 - 4168966 CGUUCUUAAUCUCAUUUUAGAGGCCAUUGACAAGAUCAAUAUAAUCACUUCACAGGGAUUUUUGGCCAGUUUUCUGGCUGGCGACGAGACUGGCCAGAGUUGGGAUCUCAUUUCCACAUA .((((((..((((......))))..(((((.....))))).............))))))((((((((((((((.(.(....).).)))))))))))))).(((((......))))).... ( -31.30, z-score = -1.13, R) >droVir3.scaffold_12875 1740639 120 + 20611582 CGUGCUCAAUCUCAUACUGGAAGCCAUUGACAAGAUAAACAUUAUAACCUCGCAAGGCUUUUUAGCCAGUUUUCUGGCCGGCGACGAGACCGGCCAGAGUUGGGAUUUAAUCUCCACGUA ((((...(((((((.((((((((((.(((.(.((..............)).))))))))).....))))).(((((((((((.....).)))))))))).))))))).......)))).. ( -38.54, z-score = -2.90, R) >droMoj3.scaffold_6496 13115266 120 - 26866924 CGUGCUCAAUCUCAUUUUGGAGGCCAUUGACAAAAUUAAUAUAAUUACAUCGCAGGGCUUCUUGGCCAGCUUCCUGGCCGGCGACGAGACUGGUCAGAGCUGGGAUCUAAUCUCAACUUA ..((..((.((((.....(((((((..(((...(((((...)))))...)))...)))))))(((((((....))))))).....)))).))..))(((.((((((...)))))).))). ( -35.00, z-score = -1.20, R) >droGri2.scaffold_15112 3830940 120 + 5172618 UGUGCUCAAUCUAAUACUGGAGGCCAUUGACAAGAUCAACAUAAUCACCUCGCAGGGUUUUCUGGCCAGUUUUCUGGCCGGCGACGAGACCGGCCAGAGUUGGGAUCUGAUCUCGACGUA .........((((....))))(((((..(((..(((.......))).((.....)))))...))))).....((((((((((.....).)))))))))((((((((...))))))))... ( -40.20, z-score = -1.36, R) >anoGam1.chr3L 8418577 120 + 41284009 CGUACUGAAUCUGAUCCUCGAAGCGAUCGACAAGAUCAACGUCAUCUCGUCGCAGGGCUUUUUGGCGUCGUUCCUGGCGAGCGACGAAAGCGGACAGAGCUGGGACAUGAUCUCCGGCUA .........((((.(((.....((((.(((...(((....)))...)))))))...(((((....((((((((.....))))))))))))))))))))((((((........)))))).. ( -40.20, z-score = 0.05, R) >apiMel3.Group2 3451136 117 + 14465785 UAUACUGAAUCUGAUCUUGGAAGCCAUCGACAAGAUCAACGUGAUCACGUCUCAAGGUUUUCU--CGUCGCUCUUU-CGGGUGACGAGAGCGGUCAAGCCUGGGACCAAAUAUCCGGAUA ........(((((((((((...........)))))))).((.(((...((((((.((((((((--((((((((...-.)))))))))))......))))))))))).....)))))))). ( -40.80, z-score = -2.41, R) >triCas2.ChLG3 11669551 108 - 32080666 AAUUUUGAAUUUAAUUUUAGAAGCAAUAGACAAGAUAAAUGUGAUAACGUCGCAAGGCUUCUUAGCCGGACUU---GCUGGCGAC---------CAAAAUUGGGAAAUGAUUUCGGGUUA ..(((((((......)))))))((..(((.((((.....((((((...)))))).((((....))))...)))---))))))(((---------(.((((((.....))))))..)))). ( -23.30, z-score = -0.68, R) >consensus CGUGCUCAAUCUGAUCCUGGAGGCCAUUGACAAGAUCAACAUAAUCACCUCGCAGGGCUUCCUGGCCAGCUUCCUGGCCGGCGACGAGACCGGCCAGAGCUGGGAUCUCAUCUCCACCUA ..................(((((((..(((...(((.......)))...)))...)))))))...(((((((....((((..........))))..)))))))................. (-18.72 = -18.42 + -0.30)

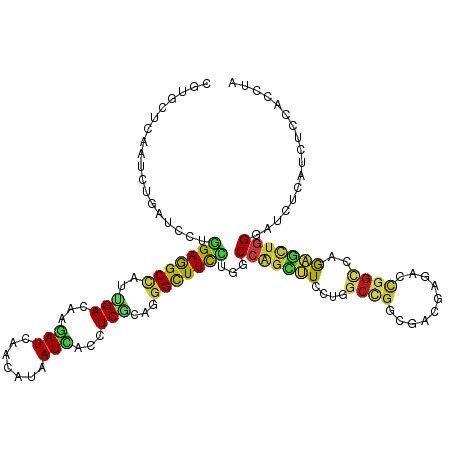
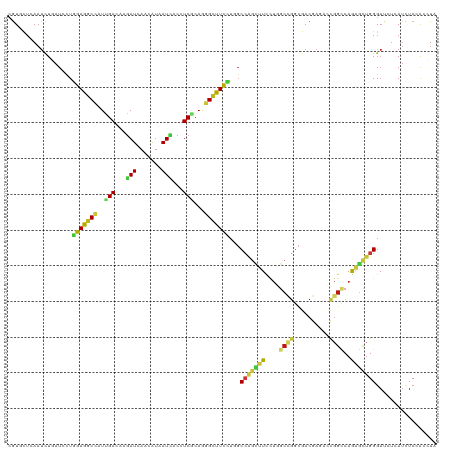
| Location | 4,755,977 – 4,756,071 |
|---|---|
| Length | 94 |
| Sequences | 14 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 80.54 |
| Shannon entropy | 0.44240 |
| G+C content | 0.55285 |
| Mean single sequence MFE | -31.44 |
| Consensus MFE | -13.16 |
| Energy contribution | -13.09 |
| Covariance contribution | -0.07 |
| Combinations/Pair | 1.67 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.546668 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 4755977 94 + 21146708 AUAAUCACCUCGCAGGGCUUCCUAGCCAGCUUCCUGGCCGGCGACGAGACCGGCCAGAGCUGGGAUCUCAUCUCCACCUAUUUGUACCAACUCU .............((((((....)))(((((..(((((((((.....).)))))))))))))(((.......))).)))............... ( -30.10, z-score = -1.39, R) >droSim1.chr2R 3433120 94 + 19596830 AUAAUCACCUCGCAGGGCUUCCUCGCCAGCGUGGUGGCCGGCGACGAGACCGGCCAGAGCUGGGAUCUCAUUUCCACCUAUCUGUACCAACUCU .....((((.(((..(((......))).))).))))((((((.....).)))))((((..(((((......)))))....)))).......... ( -33.70, z-score = -1.46, R) >droSec1.super_1 2393775 94 + 14215200 AUAAUCACCUCGCAGGGCUUCCUCGCCAGCUUCCUGGCCGGCGACGAGACCGGCCAGAGCUGGGAUCUCAUUUCCACCUAUCUGUACCAACUCU ...........(((((((......)))((((..(((((((((.....).))))))))))))((((......))))......))))......... ( -32.40, z-score = -2.17, R) >droYak2.chr2L 17414538 94 + 22324452 AUUAUCACCUCGCAGGGCUUUCUCGCCAGCUUUCUGGCUGGCGACGAAACCGGCCAGAGCUGGGAUCUCAUCUCCACCUAUCUCUACCAACUCU ...............((.(((((((((((((....))))))))).))))))((..((((.((((............)))).)))).))...... ( -31.40, z-score = -3.12, R) >droEre2.scaffold_4929 8745672 94 - 26641161 AUAAUCACCUCGCAGGGCUUCCUCGCCAGCUUCCUGGCCGGCGACGAGACCGGUCAGAGCUGGGAUCUCAUCUCCACCUAUCUUUACCAACUCC ...(((....((.(((....)))))((((((..(((((((((.....).)))))))))))))))))............................ ( -27.60, z-score = -1.08, R) >droAna3.scaffold_13266 18620906 94 - 19884421 AUAAUUACAUCUCAGGGCUUCCUGGCCAGCUUCCUGGCCGGAGACGAGACUGGCCAAAGCUGGGACCUCAUCUCCACCUACCUGUACCAACUAC ............((((.......((((((....))))))(((((.(((.(..((....))..)...))).))))).....)))).......... ( -33.40, z-score = -2.94, R) >dp4.chr3 13904219 94 + 19779522 AUUAUCACGUCGCAGGGCUUCCUGGCCAGCUUUCUCGCUGGCGACGAGACCGGCCAAAGCUGGGACCUCAUCUCCACGUACCUCUAUCAACUGC ......(((((.((((....))))((((((......)))))))))(((.(((((....)))))...)))........))............... ( -31.20, z-score = -1.82, R) >droPer1.super_4 5747925 94 - 7162766 AUCAUCACGUCGCAGGGCUUCCUGGCCAGCUUUCUCGCUGGCGACGAGACCGGCCAAAGCUGGGACCUCAUCUCCACGUACCUCUAUCAACUGC ......(((((.((((....))))((((((......)))))))))(((.(((((....)))))...)))........))............... ( -31.20, z-score = -1.74, R) >droWil1.scaffold_180697 1435958 94 - 4168966 AUAAUCACUUCACAGGGAUUUUUGGCCAGUUUUCUGGCUGGCGACGAGACUGGCCAGAGUUGGGAUCUCAUUUCCACAUAUUUGUAUCAACUAC ..((((.((.....))))))(((((((((((((.(.(....).).)))))))))))))(((((((.......)))(((....)))..))))... ( -27.30, z-score = -2.34, R) >droVir3.scaffold_12875 1740679 94 + 20611582 AUUAUAACCUCGCAAGGCUUUUUAGCCAGUUUUCUGGCCGGCGACGAGACCGGCCAGAGUUGGGAUUUAAUCUCCACGUACCUGUACCAACUGC ...........(((.((((....)))).....((((((((((.....).)))))))))(((((((.......)))..(((....)))))))))) ( -32.00, z-score = -2.84, R) >droMoj3.scaffold_6496 13115306 94 - 26866924 AUAAUUACAUCGCAGGGCUUCUUGGCCAGCUUCCUGGCCGGCGACGAGACUGGUCAGAGCUGGGAUCUAAUCUCAACUUAUCUCUACCAGCUAC .......(.(((((((....)))((((((....)))))).)))).)((.(((((.((((.((((((...))))))......))))))))))).. ( -29.30, z-score = -1.57, R) >droGri2.scaffold_15112 3830980 94 + 5172618 AUAAUCACCUCGCAGGGUUUUCUGGCCAGUUUUCUGGCCGGCGACGAGACCGGCCAGAGUUGGGAUCUGAUCUCGACGUAUUUGUAUCAACUGC ...........((((((((....)))).....((((((((((.....).)))))))))((((((((...)))))))).............)))) ( -34.60, z-score = -2.29, R) >anoGam1.chr3L 8418617 94 + 41284009 GUCAUCUCGUCGCAGGGCUUUUUGGCGUCGUUCCUGGCGAGCGACGAAAGCGGACAGAGCUGGGACAUGAUCUCCGGCUAUCUGUAUCAGCUGC ......(((((((...(((....))).((((.....))))))))))).(((..(((((((((((........))))))..)))))....))).. ( -34.70, z-score = -0.73, R) >apiMel3.Group2 3451176 91 + 14465785 GUGAUCACGUCUCAAGGUUUUCU--CGUCGCUCUUU-CGGGUGACGAGAGCGGUCAAGCCUGGGACCAAAUAUCCGGAUACCUGUAUCAACUGU ........((((((.((((((((--((((((((...-.)))))))))))......)))))))))))..........((((....))))...... ( -31.20, z-score = -2.03, R) >consensus AUAAUCACCUCGCAGGGCUUCCUGGCCAGCUUCCUGGCCGGCGACGAGACCGGCCAGAGCUGGGAUCUCAUCUCCACCUAUCUGUACCAACUGC .........................(((((((....(((((........)))))..)))))))............................... (-13.16 = -13.09 + -0.07)



Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:09:09 2011