
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 4,752,131 – 4,752,222 |
| Length | 91 |
| Max. P | 0.718916 |

| Location | 4,752,131 – 4,752,222 |
|---|---|
| Length | 91 |
| Sequences | 9 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 63.75 |
| Shannon entropy | 0.68760 |
| G+C content | 0.51683 |
| Mean single sequence MFE | -31.87 |
| Consensus MFE | -11.51 |
| Energy contribution | -11.23 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.53 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.718916 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 4752131 91 + 21146708 UGUUCCAUGCCCAAAAGG-AUGCAGAUGGA-----------------GCUGGCGAGU-CGAAUGUGUC------GGCAAAUGCCAAUUGGUAUUGCGGUGGCUUGCCGGUCGGAAU .(((((.(((((....))-..)))....((-----------------.(((((((((-(((.....))------.((((.((((....))))))))...))))))))))))))))) ( -33.70, z-score = -1.50, R) >droVir3.scaffold_12875 1737524 73 + 20611582 ----------------------------------------CUGGUCAUUCAGGGCGCUUGGGUAAACC--AAUUGGCAAUUGGCAAUUGGUAUUUGGAUGACAUGCAUGCUGUCU- ----------------------------------------...((((((((((((......))..(((--(((((.(....).)))))))).)))))))))).............- ( -22.20, z-score = -1.43, R) >droPer1.super_4 5745012 94 - 7162766 ---UCUGGCAUUGGAACUUGUCCCUAAAGG-----------------GCUCACGUGUAUGUGUGUGUG--AGGGAGAAAAUACCAAUUGGUAUUGUGGUUGCUUGCCGGUUGGAAU ---.((((((..(.(((((.(((((...(.-----------------((.((((....)))).)).).--))))).))((((((....))))))...))).).))))))....... ( -31.70, z-score = -2.66, R) >dp4.chr3 13901299 96 + 19779522 ---UCUGGCAUUGGAACUUGUCCCCAAAGG-----------------GCUCACGUGUAUGUGUGUGUGUGAGGGGGAAAAUACCAAUUGGUAUUGUGGUUGCUUGCCGGUUGGAAU ---.((((((..(.((((..(((((.....-----------------.((((((..((....))..))))))))))).((((((....))))))..)))).).))))))....... ( -34.60, z-score = -2.52, R) >droAna3.scaffold_13266 18617528 109 - 19884421 AUCUCUAGACCCAAAAGGGAUGCAGACGGCCAUGGACUUGGACUUUCACCGGGUAGC-CGAAUGUGUU------GCCAAAUACCAAUUGGUAUUGUGGUGGCUUGCCGGUCGGAAU ...(((.((((..........(((..((((.....((((((.......)))))).))-))..)))((.------.(((((((((....)))))).)))..)).....))))))).. ( -36.30, z-score = -0.83, R) >droEre2.scaffold_4929 8741919 91 - 26641161 UUCUCCAUGCCCACAAGG-AUGCAGAUGGA-----------------GCUGGCGAGU-CGAAUGUGUC------GGCAAAUGCCAAUUGGUAUUGCGGUGGCUUGCCGGUCGGAAU ..((((((((((....))-..)))..))))-----------------)(((((((((-(((.....))------.((((.((((....))))))))...))))))))))....... ( -32.90, z-score = -0.86, R) >droYak2.chr2L 17410668 91 + 22324452 UGCUCUACGCCCAAAAGG-AUGCAGAUGGA-----------------GCUGGCGAGU-CGAAUGUGUC------GGCAAAUGCCAAUUGGUAUUACGGUGGCUUGUCGGUCGGAAU .(((((((((((....))-..)).).))))-----------------))((((((((-(.........------(((....))).((((......)))))))))))))........ ( -26.40, z-score = 0.18, R) >droSec1.super_1 2390019 91 + 14215200 UGCUCCAUGCCCAAAAGG-AUGCAGAUGGA-----------------GCUGGCGAGU-CGAAUGUGUC------GGCAAAUGCCAAUUGGUAUUGCGGUGGCUUGCCGGUCGGAAU .(((((((((((....))-..)))..))))-----------------))((((((((-(((.....))------.((((.((((....))))))))...)))))))))........ ( -34.50, z-score = -1.42, R) >droSim1.chr2R 3429428 91 + 19596830 UGCUCCAUGCCCAAAAGG-AUGCAGAUGGA-----------------GCUGGCGAGU-CGAAUGUGUC------GGCAAAUGCCAAUUGGUAUUGCGGUGGCUUGCCGGUCGGAAU .(((((((((((....))-..)))..))))-----------------))((((((((-(((.....))------.((((.((((....))))))))...)))))))))........ ( -34.50, z-score = -1.42, R) >consensus UGCUCCAUGCCCAAAAGG_AUGCAGAUGGA_________________GCUGGCGAGU_CGAAUGUGUC______GGCAAAUGCCAAUUGGUAUUGCGGUGGCUUGCCGGUCGGAAU ................................................(((((.........................((((((....)))))).((((.....)))))))))... (-11.51 = -11.23 + -0.28)
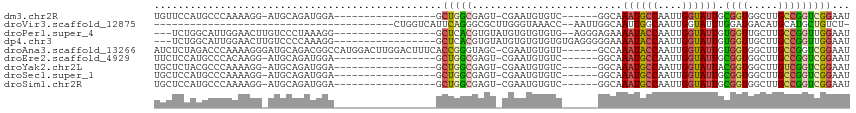

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:09:06 2011