| Sequence ID | dm3.chr2R |
|---|---|
| Location | 4,739,725 – 4,739,818 |
| Length | 93 |
| Max. P | 0.522467 |

| Location | 4,739,725 – 4,739,818 |
|---|---|
| Length | 93 |
| Sequences | 12 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 73.58 |
| Shannon entropy | 0.57918 |
| G+C content | 0.62152 |
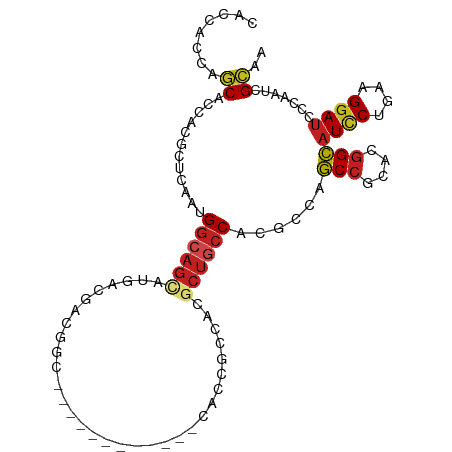
| Mean single sequence MFE | -27.49 |
| Consensus MFE | -13.08 |
| Energy contribution | -13.47 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.27 |
| Mean z-score | -0.99 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.522467 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
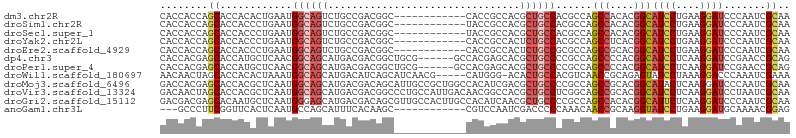
>dm3.chr2R 4739725 93 + 21146708 CACCACCAGCACCACACUGAAUGGCAGUCUGCCGACGGC------------CACCGCCACGCUGCCACGCCAGCCACACGGCAUCCUGAAGGAUCCCAAUCGCAA ........((......(((..(((((((.((.((.....------------...)).)).)))))))...)))......((.((((....)))).))....)).. ( -25.70, z-score = -0.76, R) >droSim1.chr2R 3416532 93 + 19596830 CACCACCAGCACCACCCUGAAUGGCAGUCUGCCGACGGC------------UACCGCCACGCUGCCACGCCAGCCACACGGCAUCCUGAAGGAUCCCAAUCGCAA ........((......(((..(((((((.((.((.....------------...)).)).)))))))...)))......((.((((....)))).))....)).. ( -25.50, z-score = -0.75, R) >droSec1.super_1 2377529 93 + 14215200 CACCACCAGCACCACCCUGAAUGGCAGUCUGCCGACGGC------------UACCGCCACGCUGCCACGCCAGCCACACGGCAUCCUGAAGGAUCCCAAUCGCAA ........((......(((..(((((((.((.((.....------------...)).)).)))))))...)))......((.((((....)))).))....)).. ( -25.50, z-score = -0.75, R) >droYak2.chr2L 17397769 93 + 22324452 CACCACCAGCACCACCCUGAAUGGCAGUCUGCCGACGGC------------CACCGCCACUCUGCCACGCCAGCCUCACGGCAUCCUGAAGGAUCCCAAUCGCAA ........((......(((..((((((((....)))(((------------....)))....)))))...)))......((.((((....)))).))....)).. ( -24.70, z-score = -0.79, R) >droEre2.scaffold_4929 8729645 93 - 26641161 CACCACCAGCACCACCCUGAAUGGCAGUCUGCCGACGGC------------CACCGCCACUCUGCCGCGCCAGCCGCACGGCAUCCUGAAGGAUCCCAAUCGCAA ........((.(((.......)))(((..(((((..(((------------....)))....(((.((....)).))))))))..))).............)).. ( -25.50, z-score = -0.12, R) >dp4.chr3 13889325 99 + 19779522 CACCACGAGCACCAUGCUCAACGGCAGCAUGACGACGGCUGCG------GCCACGAGCACGCUGCCCCGCCAGCCCCACGGCAUCCUCAAGGAUCCGAACCGCAG ......((((.....))))...((((((.((.((..(((....------))).))..)).))))))......((....(((.((((....)))))))....)).. ( -34.20, z-score = -1.66, R) >droPer1.super_4 5732545 99 - 7162766 CACCACGAGCACCAUGCUCAACGGCAGCAUGACGACGGCUGCG------GCCACGAGCACGCUGCCCCGCCAGCCCCACGGCAUCCUCAAGGAUCCGAACCGCAG ......((((.....))))...((((((.((.((..(((....------))).))..)).))))))......((....(((.((((....)))))))....)).. ( -34.20, z-score = -1.66, R) >droWil1.scaffold_180697 1417715 99 - 4168966 AACAACUAGCACCACACUAAAUGGCAGCAUGACAUCAGCAUCAACG-----CAUGGG-ACACUGCCACGUCAACCGCAGAGUAUCCUAAAGGACCCAAAUCGAAA ........((.(((.......)))..((.((....))))......)-----).((((-((.((((..........)))).)).(((....)))))))........ ( -18.20, z-score = -0.42, R) >droMoj3.scaffold_6496 13098055 105 - 26866924 GACCACGAGCACCACGCUCAAUGGCAGCAUGACGACAGCAUUGCCGCUGGCCACAUCGACGCUGCCCCGCCAGCCGCACGGCAUACUCAAGGAUCCCAAUCGCAA ..((..((((.....))))...(((((((((....((((......))))....)))....))))))......(((....)))........))............. ( -29.30, z-score = -0.17, R) >droVir3.scaffold_13324 2949395 105 + 2960039 GACAACUAGCACCACGCUCAAUGGCAGCAUGACGACGGCCCUGCCAUUGACAACGGCCACGCUGCCUCGGCAGCCGCACGGCAUCCUCAAGGAUCCUAAUCGCAA ........((.....((((((((((((..((....))...))))))))))..........(((((....))))).))..((.((((....)))))).....)).. ( -35.30, z-score = -1.95, R) >droGri2.scaffold_15112 3813768 105 + 5172618 GACGACGAGCACAAUGCUCAAUGGGAGCAUGACGACAGCGUUGCCACUUGCCACAUCAACGCUGCCCCGCCAGCCACACGGCAUUCUCAAGGAUCCCAAUCGCAA (.(((.((((.....))))..(((((.(.(((.((((((((((.............))))))))....(((........)))..))))).)..))))).)))).. ( -32.12, z-score = -1.76, R) >anoGam1.chr3L 8388271 90 + 41284009 ---GCCCUUCGGUUCACUCAAUGCCAGCAUUUCACAAGC------------CGUCCAAUCGACCCCCAAACAACCGCAAGGUAUCCUGAAGGAUGCAAAACGGAG ---(((((((((((.....((((....)))).....)))------------)(((.....))).........(((....))).....)))))..))......... ( -19.70, z-score = -1.07, R) >consensus CACCACCAGCACCACGCUCAAUGGCAGCAUGACGACGGC____________CACCGCCACGCUGCCACGCCAGCCGCACGGCAUCCUGAAGGAUCCCAAUCGCAA ........((............((((((................................))))))......(((....)))((((....)))).......)).. (-13.08 = -13.47 + 0.39)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:09:04 2011