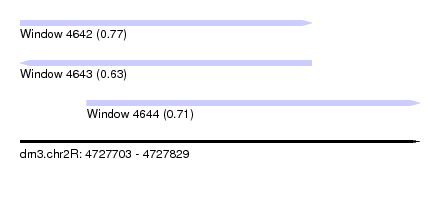
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 4,727,703 – 4,727,829 |
| Length | 126 |
| Max. P | 0.767280 |

| Location | 4,727,703 – 4,727,795 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 84.62 |
| Shannon entropy | 0.28161 |
| G+C content | 0.51665 |
| Mean single sequence MFE | -32.95 |
| Consensus MFE | -22.00 |
| Energy contribution | -23.87 |
| Covariance contribution | 1.86 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.767280 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
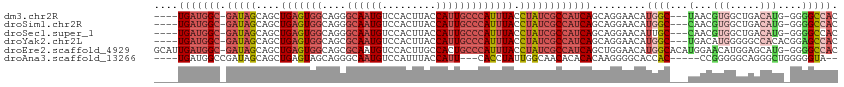
>dm3.chr2R 4727703 92 + 21146708 ---------CGAGGACGAUGAUGAUGGCAAUGAUGGC-GAUAGCAGCUGAGUGGCAGGGCAAUGUCCACUUACCAUUGCCCAUUUACCUAUCGCCAUCAGCA ---------.................((..(((((((-(((((..(((....))).((((((((.........))))))))......)))))))))))))). ( -36.80, z-score = -3.20, R) >droSim1.chr2R 3404581 92 + 19596830 ---------CGAGGACGAUGAUGAUGGCAAUGAUGGC-GAUAGCAGCUGAGUGGCAGGGCAAUGUCCACUUACCAUUGCCCAUUUACCUAUCGCCAUCAGCA ---------.................((..(((((((-(((((..(((....))).((((((((.........))))))))......)))))))))))))). ( -36.80, z-score = -3.20, R) >droSec1.super_1 2365511 92 + 14215200 ---------CGAGGACGAUGAUGAUGGCAAUGAUGGC-GAUAGCAGCUGAGUGGCAGGGCAAUGUCCACUUACCAUUGCCCAUUUACCUAUCGCCAUCAGCA ---------.................((..(((((((-(((((..(((....))).((((((((.........))))))))......)))))))))))))). ( -36.80, z-score = -3.20, R) >droYak2.chr2L 17385995 92 + 22324452 ---------CAAAGACGAUGAUGAUGGCAAUGAUGGC-GAUAGCAGCUGAGUGGCAGCGCAAUGUCCACUUACCAUUGCCCAUUUACCUAUCGCCAUCAGCA ---------.................((..(((((((-(((((..((((.....))))((((((.........))))))........)))))))))))))). ( -32.10, z-score = -2.61, R) >droEre2.scaffold_4929 8717895 101 - 26641161 CGAGGACGAUGAUGAUGGCAAUGAUGGCAUUGAUGGC-GAUAGCAGCUGAGUGGCAGCGCAAUGUCCACUUGCCACUGCCCAUUUACCUAUCGCCAUCAGCU .............((((.((....)).))))((((((-(((((....((((((((((.((((.(....)))))..))).))))))).))))))))))).... ( -35.40, z-score = -1.17, R) >droAna3.scaffold_13266 18593946 90 - 19884421 ---------CCGGGUCGAUGAUGAUGGCAAUGAUGGCCGAUAGCAGCUGAGUAGCAGGGCAAUGUCCAUUUACCAUUCACCUAUUGGCAACACACACAA--- ---------....(((((((.((((((....(((((.((...((..(((.....))).))..)).)))))..))).)))..)))))))...........--- ( -19.80, z-score = 1.36, R) >consensus _________CGAGGACGAUGAUGAUGGCAAUGAUGGC_GAUAGCAGCUGAGUGGCAGGGCAAUGUCCACUUACCAUUGCCCAUUUACCUAUCGCCAUCAGCA ..............................(((((((.(((((..(((....))).((((((((.........))))))))......))))))))))))... (-22.00 = -23.87 + 1.86)

| Location | 4,727,703 – 4,727,795 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 84.62 |
| Shannon entropy | 0.28161 |
| G+C content | 0.51665 |
| Mean single sequence MFE | -32.48 |
| Consensus MFE | -22.91 |
| Energy contribution | -23.92 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.633982 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 4727703 92 - 21146708 UGCUGAUGGCGAUAGGUAAAUGGGCAAUGGUAAGUGGACAUUGCCCUGCCACUCAGCUGCUAUC-GCCAUCAUUGCCAUCAUCAUCGUCCUCG--------- .((((((((((((((......((((((((.........)))))))).((......))..)))))-)))))))..)).................--------- ( -35.10, z-score = -2.70, R) >droSim1.chr2R 3404581 92 - 19596830 UGCUGAUGGCGAUAGGUAAAUGGGCAAUGGUAAGUGGACAUUGCCCUGCCACUCAGCUGCUAUC-GCCAUCAUUGCCAUCAUCAUCGUCCUCG--------- .((((((((((((((......((((((((.........)))))))).((......))..)))))-)))))))..)).................--------- ( -35.10, z-score = -2.70, R) >droSec1.super_1 2365511 92 - 14215200 UGCUGAUGGCGAUAGGUAAAUGGGCAAUGGUAAGUGGACAUUGCCCUGCCACUCAGCUGCUAUC-GCCAUCAUUGCCAUCAUCAUCGUCCUCG--------- .((((((((((((((......((((((((.........)))))))).((......))..)))))-)))))))..)).................--------- ( -35.10, z-score = -2.70, R) >droYak2.chr2L 17385995 92 - 22324452 UGCUGAUGGCGAUAGGUAAAUGGGCAAUGGUAAGUGGACAUUGCGCUGCCACUCAGCUGCUAUC-GCCAUCAUUGCCAUCAUCAUCGUCUUUG--------- .((((((((((((((........((((((.........))))))((((.....))))..)))))-)))))))..)).................--------- ( -32.80, z-score = -1.87, R) >droEre2.scaffold_4929 8717895 101 + 26641161 AGCUGAUGGCGAUAGGUAAAUGGGCAGUGGCAAGUGGACAUUGCGCUGCCACUCAGCUGCUAUC-GCCAUCAAUGCCAUCAUUGCCAUCAUCAUCGUCCUCG .(.((((((((((.((((....(((.(((((((((((.((......)))))))....)))))).-))).....))))...)))))))))).).......... ( -37.20, z-score = -1.69, R) >droAna3.scaffold_13266 18593946 90 + 19884421 ---UUGUGUGUGUUGCCAAUAGGUGAAUGGUAAAUGGACAUUGCCCUGCUACUCAGCUGCUAUCGGCCAUCAUUGCCAUCAUCAUCGACCCGG--------- ---(((.((.....)))))..(((((.((((((((((.....((...(((....))).))......))))..)))))))))))..........--------- ( -19.60, z-score = 1.49, R) >consensus UGCUGAUGGCGAUAGGUAAAUGGGCAAUGGUAAGUGGACAUUGCCCUGCCACUCAGCUGCUAUC_GCCAUCAUUGCCAUCAUCAUCGUCCUCG_________ ...((((((((((.((((...((((((((.........)))))))))))).....((........))....))))))))))..................... (-22.91 = -23.92 + 1.00)
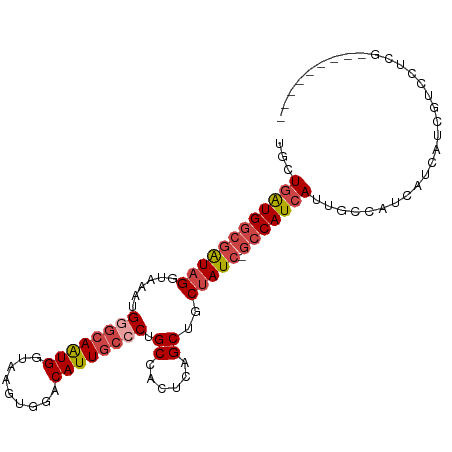
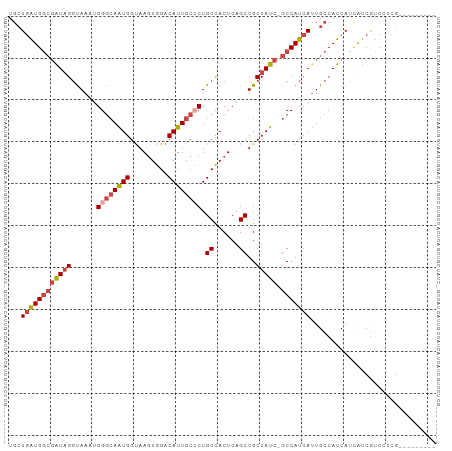
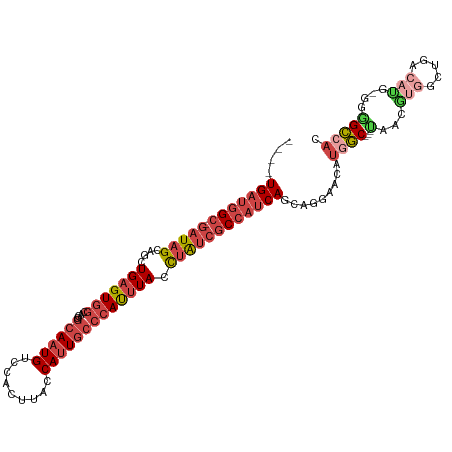
| Location | 4,727,724 – 4,727,829 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 78.45 |
| Shannon entropy | 0.39797 |
| G+C content | 0.56253 |
| Mean single sequence MFE | -42.72 |
| Consensus MFE | -18.88 |
| Energy contribution | -21.47 |
| Covariance contribution | 2.59 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.707517 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 4727724 105 + 21146708 ----UGAUGGC-GAUAGCAGCUGAGUGGCAGGGCAAUGUCCACUUACCAUUGCCCAUUUACCUAUCGCCAUCAGCAGGAACAUGGC---UAACGUGGCUGACAUG-GGGGCCAC ----(((((((-(((((..(((....))).((((((((.........))))))))......)))))))))))).........((((---(..((((.....))))-..))))). ( -49.10, z-score = -4.28, R) >droSim1.chr2R 3404602 105 + 19596830 ----UGAUGGC-GAUAGCAGCUGAGUGGCAGGGCAAUGUCCACUUACCAUUGCCCAUUUACCUAUCGCCAUCAGCAGGAACAUGGC---CAACGUGGCUGACAUG-GGGGCCAC ----(((((((-(((((..(((....))).((((((((.........))))))))......)))))))))))).........((((---(..((((.....))))-..))))). ( -51.00, z-score = -4.55, R) >droSec1.super_1 2365532 105 + 14215200 ----UGAUGGC-GAUAGCAGCUGAGUGGCAGGGCAAUGUCCACUUACCAUUGCCCAUUUACCUAUCGCCAUCAGCAGGAACAUUGC---CAACGUGGCUGACAUG-GGGGCCAC ----(((((((-(((((..(((....))).((((((((.........))))))))......))))))))))))...........((---(..((((.....))))-..)))... ( -46.10, z-score = -3.48, R) >droYak2.chr2L 17386016 106 + 22324452 ----UGAUGGC-GAUAGCAGCUGAGUGGCAGCGCAAUGUCCACUUACCAUUGCCCAUUUACCUAUCGCCAUCAGCAGGAACAUGGC---UGACAUGGGGGCCACACGGAGCCAC ----(((((((-(((((..((((.....))))((((((.........))))))........)))))))))))).......((((..---...))))((..((....))..)).. ( -38.90, z-score = -1.64, R) >droEre2.scaffold_4929 8717921 112 - 26641161 GCAUUGAUGGC-GAUAGCAGCUGAGUGGCAGCGCAAUGUCCACUUGCCACUGCCCAUUUACCUAUCGCCAUCAGCUGGAACAUGGCACAUGGAACAUGGAGCAUG-GGGGCCAC .((((((((((-(((((....((((((((((.((((.(....)))))..))).))))))).))))))))))))).)).....((((.((((..........))))-...)))). ( -42.60, z-score = -1.20, R) >droAna3.scaffold_13266 18593967 100 - 19884421 ----UGAUGGCCGAUAGCAGCUGAGUAGCAGGGCAAUGUCCAUUUACCAUU---CACCUAUUGGCAACACACACAAGGGGCACCAC-----CCGGGGGCAGGGCUGGGGGUA-- ----.....((((((((..(((....))).((((...))))..........---...))))))))................(((.(-----((((........)))))))).-- ( -28.60, z-score = 1.52, R) >consensus ____UGAUGGC_GAUAGCAGCUGAGUGGCAGGGCAAUGUCCACUUACCAUUGCCCAUUUACCUAUCGCCAUCAGCAGGAACAUGGC___UAACGUGGCUGACAUG_GGGGCCAC ....(((((((.(((((....(((((((....((((((.........))))))))))))).)))))))))))).........((((.......(((.....))).....)))). (-18.88 = -21.47 + 2.59)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:09:03 2011