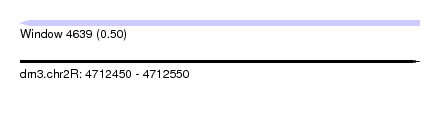
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 4,712,450 – 4,712,550 |
| Length | 100 |
| Max. P | 0.500000 |

| Location | 4,712,450 – 4,712,550 |
|---|---|
| Length | 100 |
| Sequences | 8 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 73.67 |
| Shannon entropy | 0.52902 |
| G+C content | 0.53556 |
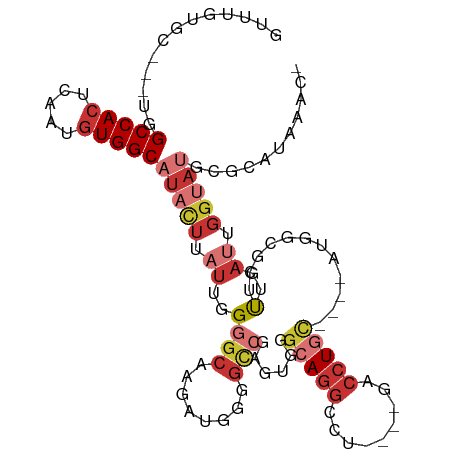
| Mean single sequence MFE | -35.28 |
| Consensus MFE | -14.65 |
| Energy contribution | -17.04 |
| Covariance contribution | 2.39 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
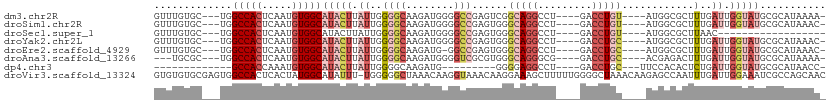
>dm3.chr2R 4712450 100 - 21146708 GUUUGUGC---UGGCCACUCAAUGUGGCAUACUUAUUGGGGCAAGAUGGGGCCGAGUCGGCAGGCCU----GACCUGU----AUGGCGCUUUGAUUGGUAUGCGCAUAAAA- .(((((((---..(((((.....)))))(((((.((..((((......(((((.........)))))----(.((...----..)))))))..)).)))))..))))))).- ( -38.60, z-score = -1.68, R) >droSim1.chr2R 3388971 100 - 19596830 GUUUGUGC---UGGCCACUCAAUGUGGCAUACUUAUUGGGGCAAGAUGGGGCCGAGUGGGCAGGCCU----GACCUGU----AUGGCGCUUUGAUUGGUAUGCGCAUAAAC- ((((((((---..(((((.....)))))(((((.((..((((......(((((.........)))))----(.((...----..)))))))..)).)))))..))))))))- ( -40.30, z-score = -2.28, R) >droSec1.super_1 2350162 83 - 14215200 GUUUGUGC---UGGCCACUCAAUGUGGCAUACUUAUUGGGGCAAGAUGGGGCCGAGUGGGCAGGCCU----GACCUGU----AUGGCGCUUAAC------------------ ....((((---(.(((((.....)))))...((((((.((.(.......).)).))))))((((...----..)))).----..))))).....------------------ ( -27.30, z-score = 0.19, R) >droYak2.chr2L 17370521 100 - 22324452 GUUUGUGC---UGGCCACUCAAUGUGGCAUACUUAUUGGGGCAAGAUGGGGCCGAGUGGGCAGGCCU----GACCUGC----AUGGCGCUUUGAUUGGUAUGCGCAUAAAC- ((((((((---..(((((.....)))))(((((.((..((((........((((.(..((((....)----).))..)----.))))))))..)).)))))..))))))))- ( -40.70, z-score = -2.07, R) >droEre2.scaffold_4929 8703045 99 + 26641161 GUUUGUGC---UGGCCACUCAAUGUGGCAUACUUAUUGGGGCAAGAUG-GGCCGAGUGGGCAGGCCU----GACCUGC----AUGGCGCUUUGAUUGGUAUGCGCAUAAAC- ((((((((---..(((((.....)))))(((((.((..((((......-.((((.(..((((....)----).))..)----.))))))))..)).)))))..))))))))- ( -40.81, z-score = -2.30, R) >droAna3.scaffold_13266 18578360 97 + 19884421 ---UGCGC---UGGCCACUCAAUGUGGCAUACUUAUUGGGGCAAGAUGGGGUCGCGUGGGCAGGGCG----GACCUGC----ACGAGACUUUGAUUGGUAUGCGCAUAAAA- ---(((((---..(((((((((((.(.....).)))))))......(..((((.(((..(((((...----..)))))----))).))))..)..))))..))))).....- ( -37.20, z-score = -2.21, R) >dp4.chr3 13859795 82 - 19779522 -------------GCCACCAAAUGUGGCAUACUUAUUGGGGCAAGAUG---------GGGGAGGCCU----GACCUGC---UUCCACACUCUGAUUGGUAUGCGCAUAACC- -------------.........(((((((((((.((..(((...(.((---------(((((((...----..))).)---)))))).)))..)).))))))).))))...- ( -28.00, z-score = -1.22, R) >droVir3.scaffold_13324 2920380 111 - 2960039 GUGUGUGCGAGUGGCCACUCACUAUGGCAUAUUU-UGGGGGCUAAACAAGGUAAACAAGGAAAGCUUUUUGGGGCUAAACAAGAGCCAAUUUGAUUGGAAAUCGCCAGCAAC .(((..((((....(((.(((...((((...(((-((..((((..(.(((((...........))))).)..))))...)))))))))...))).)))...))))..))).. ( -29.30, z-score = -0.39, R) >consensus GUUUGUGC___UGGCCACUCAAUGUGGCAUACUUAUUGGGGCAAGAUGGGGCCGAGUGGGCAGGCCU____GACCUGC____AUGGCGCUUUGAUUGGUAUGCGCAUAAAC_ .............(((((.....)))))(((((.((..(((.........((((.....(((((.(.....).))))).....)))).)))..)).)))))........... (-14.65 = -17.04 + 2.39)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:08:59 2011