| Sequence ID | dm3.chr2R |
|---|---|
| Location | 4,701,567 – 4,701,695 |
| Length | 128 |
| Max. P | 0.808672 |

| Location | 4,701,567 – 4,701,695 |
|---|---|
| Length | 128 |
| Sequences | 8 |
| Columns | 140 |
| Reading direction | reverse |
| Mean pairwise identity | 73.73 |
| Shannon entropy | 0.47816 |
| G+C content | 0.46844 |
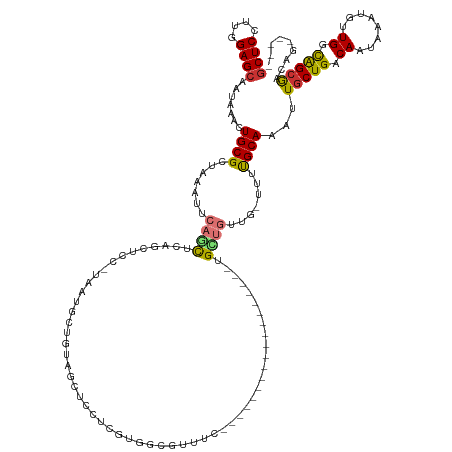
| Mean single sequence MFE | -38.38 |
| Consensus MFE | -13.56 |
| Energy contribution | -13.38 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.37 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.808672 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
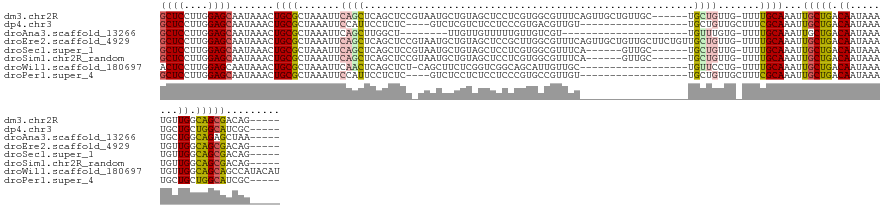
>dm3.chr2R 4701567 128 - 21146708 GCUCCUUGGAGCAAUAAACUGCGCUAAAUUCAGCUCAGCUCCGUAAUGCUGUAGCUCCUCGUGGCGUUUCAGUUGCUGUUGC------UGCUGUUG-UUUUGCAAAUUGCUGACAAUAAAUGUUGGCAGCGACAG----- ((((....))))....((((((((((.....((((((((........)))).)))).....)))))...))))).(((((((------((((((((-((..((.....)).)))))).......)))))))))))----- ( -50.11, z-score = -3.94, R) >dp4.chr3 13847045 113 - 19779522 GCUCCUUGGAGCAAUAAACUGCGCUAAAUUCCAUUCCUCUC----GUCUCGUCUCCUCCCGUGACGUUGU------------------UGCUGUUGCUUUCGCAAAUUGCUGACAAUAAAUGCUGCUGGCAUCGC----- ((((....))))..........((((.....((((......----(((.((........)).)))(((((------------------(((..((((....))))...)).)))))).))))....)))).....----- ( -26.30, z-score = -1.46, R) >droAna3.scaffold_13266 18566913 105 + 19884421 GCUCCUUGGAGCAAUAAACUGCGCUAAAUUCAGCUUGGCU--------UUGUUGUUUUUGUUGUCGU---------------------UGUUUGUG-UUUUGCAAAUUGCUGACAAUAAAUGCUGGCAGAGCUAA----- ((((....))))..........(((......))).(((((--------(((((((.(((((((((((---------------------.((((((.-....)))))).)).))))))))).)).)))))))))).----- ( -37.40, z-score = -3.92, R) >droEre2.scaffold_4929 8693070 134 + 26641161 GCUCCUUGGAGCAAUAAACUGCGCUAAAUUCAGCUCAGCUCCGUAAUGCUGUAGCUCCGCUUGGCGUUUCAGUUGCUGUUGCUUCUGUUGCUGUUG-UUUUGCAAAUUGCUGACAAUAAAUGUUGGCAGCGACAG----- ((((....))))....(((((((((((....((((((((........)))).))))....))))))...))))).(((((((.....((((.....-....))))..(((..(((.....)))..))))))))))----- ( -48.30, z-score = -2.84, R) >droSec1.super_1 2339285 122 - 14215200 GCUCCUUGGAGCAAUAAACUGCGCUAAAUUCAGCUCAGCUCCGUAAUGCUGUAGCUCCUCGUGGCGUUUCA------GUUGC------UGCUGUUG-UUUUGCAAAUUGCUGACAAUAAAUGUUGGCAGCGACAG----- ((((....))))........((((((.....((((((((........)))).)))).....))))))....------(((((------((((((((-((..((.....)).)))))).......)))))))))..----- ( -45.31, z-score = -3.30, R) >droSim1.chr2R_random 1605155 122 - 2996586 GCUCCUUGGAGCAAUAAACUGCGCUAAAUUCAGCUCAGCUCCGUAAUGCUGUAGCUCCUCGUGGCGUUUCA------GUUGC------UGCUGUUG-UUUUGCAAAUUGCUGACAAUAAAUGUUGGCAGCGACAG----- ((((....))))........((((((.....((((((((........)))).)))).....))))))....------(((((------((((((((-((..((.....)).)))))).......)))))))))..----- ( -45.31, z-score = -3.30, R) >droWil1.scaffold_180697 1366171 120 + 4168966 ACUCCUUGGAGCAAUAAACUGCGCUAAAUUCAACUCAGCUCU-CAGCUUCUCGGUCGGCAGCAUUGUUGC------------------UGUUCCUG-UUUUGCAAAUUGCUGACAAUAAAUGUUGGCAGCAGCCAUACAU .(((....))).......(((((((((.......(((((...-..((....(((..(((((((....)))------------------)))).)))-....)).....))))).........))))).))))........ ( -32.59, z-score = -0.81, R) >droPer1.super_4 5689599 113 + 7162766 GCUCCUUGGAGCAAUAAACUGCGCUAAAUUCCAUUCCUCUC----GUCUCCUCUCCUCCCGUGCCGUUGU------------------UGCUGUUGCUUUCGCAAAUUGCUGACAAUAAAUGCUGCUGGCAUCGC----- ((((....))))........((((.................----...............)))).(((((------------------(((..((((....))))...)).))))))..((((.....))))...----- ( -21.75, z-score = -0.26, R) >consensus GCUCCUUGGAGCAAUAAACUGCGCUAAAUUCAGCUCAGCUCC_UAAUGCUGUAGCUCCUCGUGGCGUUUC__________________UGCUGUUG_UUUUGCAAAUUGCUGACAAUAAAUGUUGGCAGCGACAG_____ ((((....)))).......((((.......((((.......................................................)))).......))))...(((((.((........)).)))))......... (-13.56 = -13.38 + -0.19)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:08:57 2011