
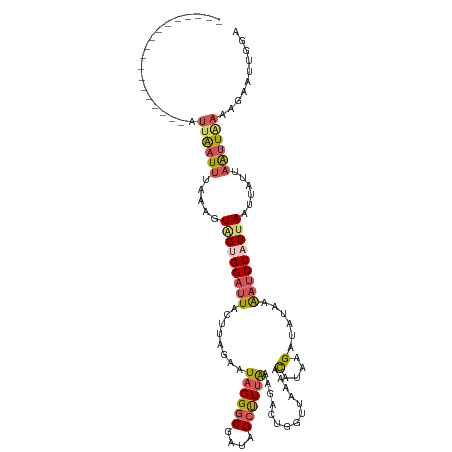
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 4,688,949 – 4,689,106 |
| Length | 157 |
| Max. P | 0.886401 |

| Location | 4,688,949 – 4,689,041 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 67.07 |
| Shannon entropy | 0.52807 |
| G+C content | 0.29060 |
| Mean single sequence MFE | -19.80 |
| Consensus MFE | -7.20 |
| Energy contribution | -8.12 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.593921 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 4688949 92 + 21146708 ------------------AUUAAUUUAAAGUAGUGGAUUACUUAGUAUAGGGCGAUAGCUUUAGAGAC---------CAUUAGCUUUUAGA-CCAUUAAUUAUUAAUUAAAAGAAUUAGA ------------------.(((((((((((((((((..(((...)))((((((....))))))....)---------)))).)))))....-...(((((.....)))))..))))))). ( -13.10, z-score = -0.18, R) >droSim1.chr2R 3370625 102 + 19596830 ------------------AUUAAUUUAAAGUAGUGGAUUACUUAGUAUAGGGCGACAGCUUUAAAGACUGGUUAAAACAUCAGAUAUAAAAUCCAUUAAUUAUUAAUUAAAAGAAUUGGA ------------------.((((((.....(((((((((........((((((....))))))....(((((......)))))......)))))))))......)))))).......... ( -20.50, z-score = -2.72, R) >droSec1.super_1 2326913 102 + 14215200 ------------------AUUAAUUUAAAGUAGUGGAUUACUUAGAAUAGGGCGACAGCUUUAAAGACUGGUUAAAACAUCAGAUAUAAAAUCCAUUAAUUAUUAAUUAAAAGAAUUGGA ------------------.((((((.....(((((((((........((((((....))))))....(((((......)))))......)))))))))......)))))).......... ( -20.50, z-score = -2.95, R) >droYak2.chr2L 17347194 120 + 22324452 GUAAAUUUAAAGUAGUGGACAGCUGGUAACUGGUGGAUUACUUAGAAUAGGGCGAUAGCUUUAAAGACUGGUUAAAACAGAAGUUAGAAGAUCCUUUAUUUACUAGCUGAAAGUAGAAUA ...................((((((((((.(((.(((((((((....((((((....))))))....(((.......))))))).....))))).))).))))))))))........... ( -29.20, z-score = -2.16, R) >droEre2.scaffold_4929 8681010 91 - 26641161 ------------------AUUAAUUUAAAGUAGGGGACUGC---AAAUAGGGCGAUGGACUGGUUAAG--------AUACAAGAUAUAAAAUCCAUCAACAAUUAAUUGAAAGCAGAGCA ------------------...........((((....))))---.......((((((((((.((....--------..)).))........))))))..(((....)))...))...... ( -15.70, z-score = -1.60, R) >consensus __________________AUUAAUUUAAAGUAGUGGAUUACUUAGAAUAGGGCGAUAGCUUUAAAGACUGGUUAAAACAUAAGAUAUAAAAUCCAUUAAUUAUUAAUUAAAAGAAUUGGA ...................((((((.....(((((((((........((((((....))))))..............(....)......)))))))))......)))))).......... ( -7.20 = -8.12 + 0.92)


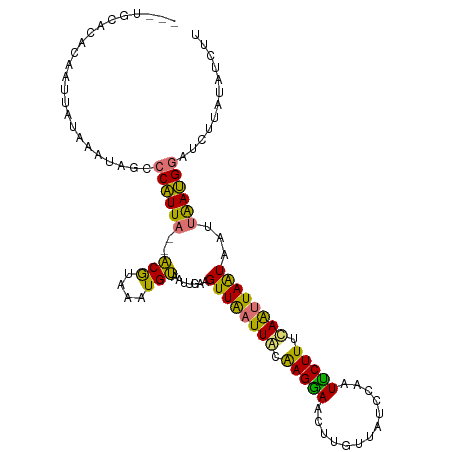
| Location | 4,689,002 – 4,689,106 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 64.06 |
| Shannon entropy | 0.58592 |
| G+C content | 0.27544 |
| Mean single sequence MFE | -17.21 |
| Consensus MFE | -10.13 |
| Energy contribution | -8.32 |
| Covariance contribution | -1.81 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.01 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.886401 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 4689002 104 - 21146708 ---UGCACACAAUUACAAAUAGCCCAUUA--ACGUAAUUGUAAUUGAAGUUGAUUACAAGGAACUUGUUAUCUAAUUCUUUUAAUUAAUAAUUAAUGG-UCUAAAAGCUA ---................(((.((((((--(..((((((.....(((...(((.(((((...))))).)))...)))...))))))....)))))))-.)))....... ( -18.20, z-score = -1.63, R) >droSim1.chr2R 3370687 108 - 19596830 AUGUGCACACAAUUAUAAAUAGCCCAUUA--ACGUAAAUGUAAUUGAAGUUGAUUACAAGGAACUUGUUAUCCAAUUCUUUUAAUUAAUAAUUAAUGGAUUUUAUAUCUG .........((..((((((....((((((--(........(((..(((...(((.(((((...))))).)))...)))..)))........)))))))..))))))..)) ( -16.99, z-score = -0.82, R) >droYak2.chr2L 17347274 110 - 22324452 CUUCAACUGCAAUUAUAAAUAGCCCUUUAACACGUAAAUGUAUUUGAAGUUAAUUCUGAGUAACUUUGUUUAUUCUACUUUCAGCUAGUAAAUAAAGGAUCUUCUAACUU ((((((.((((.((((.................)))).)))).))))))........(((...(((((((((((((......))..)))))))))))...)))....... ( -16.13, z-score = -0.56, R) >droEre2.scaffold_4929 8681061 87 + 26641161 ---------------------AUGCAUUU--GCAUUAAUGCAUUCGAAGUUAAUUGCGAGUAACUCGUUUUGCUCUGCUUUCAAUUAAUUGUUGAUGGAUUUUAUAUCUU ---------------------(((((((.--.....)))))))(((((((((((((.(((((.............))))).))))))))).)))).((((.....)))). ( -17.52, z-score = -1.02, R) >consensus ___UGCACACAAUUAUAAAUAGCCCAUUA__ACGUAAAUGUAAUUGAAGUUAAUUACAAGGAACUUGUUAUCCAAUUCUUUCAAUUAAUAAUUAAUGGAUCUUAUAUCUU .......................((((((..(((....))).......((((((((.(((((.............))))).))))))))...))))))............ (-10.13 = -8.32 + -1.81)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:08:53 2011