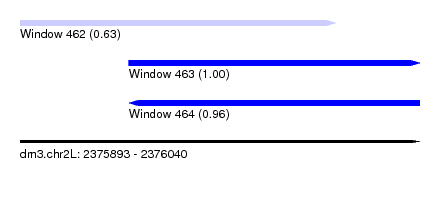
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 2,375,893 – 2,376,040 |
| Length | 147 |
| Max. P | 0.995171 |

| Location | 2,375,893 – 2,376,009 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 85.93 |
| Shannon entropy | 0.26659 |
| G+C content | 0.31927 |
| Mean single sequence MFE | -17.44 |
| Consensus MFE | -9.75 |
| Energy contribution | -11.42 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.632470 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
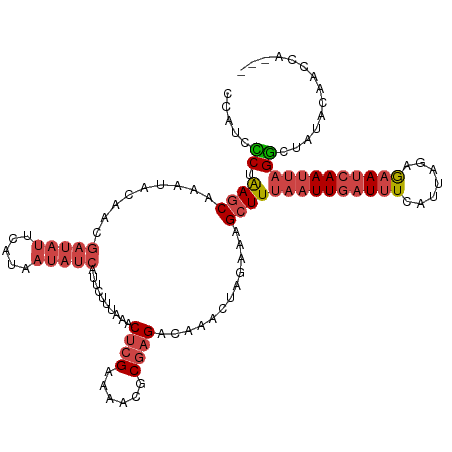
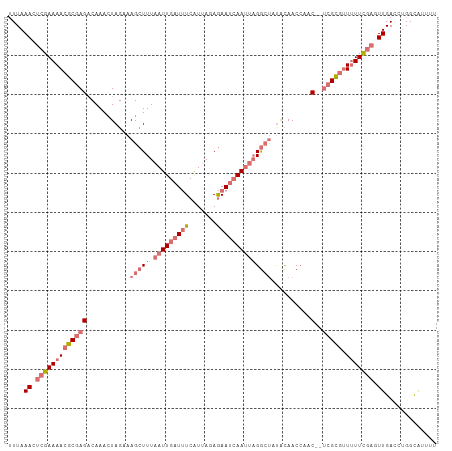
>dm3.chr2L 2375893 116 + 23011544 CCAUCCCUAAGCAAAUACAACGAUAUUCAUAAUAUCAAUCUUUAAACUCGAAAAAGCGAGACAAACUAGAAAGCUUUAAUUGAUUUCGUUACAGAAUCAAUUAGGCUAUACAACCA--- ......(((............(((((.....)))))..........((((......))))......)))..(((((.(((((((((.......)))))))))))))).........--- ( -18.80, z-score = -3.06, R) >droAna3.scaffold_12916 2101585 112 - 16180835 CCAUCUCUAACCAAACACAACUAUAUUCAUA----UGUUCAUUAAACAAGAAAACGCGAG-CAAA-UACCAAGAUUUAAUUGAUAUCAU-AGAAAACAAACGUGAAUCUGUAACAUACA .............................((----(((((..............(....)-....-.....(((((((.(((.......-......)))...)))))))).)))))).. ( -7.22, z-score = 0.92, R) >droEre2.scaffold_4929 2415338 116 + 26641161 CCAUCUCUGAGCAAAUACAACGAUAUUCAUAAUAUCAUUCUUUAAACUCGAAAACGCGAGACAAACUAGCAAGCUUUAAUUGAUUUCAUCAGAGAAUCAAUUAGGCUAUACAACCA--- ..........((.........(((((.....)))))..........((((......))))........)).(((((.(((((((((.......)))))))))))))).........--- ( -20.80, z-score = -2.96, R) >droYak2.chr2L 2358449 116 + 22324452 CCAUCUCUAAGCAAAUACAACGAUAUUCAUAAUAUCAUUCUUUAAACUCGAAAACGCGAGACAAACUAGCAAGCUUUAAUUGAUUUCAUUAGAGAAUCAAUUAGGCGACACAACCA--- ..........((.........(((((.....)))))..........((((......))))........))..((((.(((((((((.......)))))))))))))..........--- ( -20.10, z-score = -3.22, R) >droSec1.super_5 547571 116 + 5866729 UCAUCCCUAAGCAAAUACAACGAUAUUCAUAAUAUCAUUCUUUAAACUCGAAAACGCGAGACAAACUAAAAAGCUUUAAUUGAUUUCGUUAGAGAAUCAAUUAGGCUAUACAACCA--- .....................(((((.....)))))..........((((......))))...........(((((.((((((((((....))).)))))))))))).........--- ( -18.80, z-score = -2.80, R) >droSim1.chr2L 2332852 116 + 22036055 UCAUCCCUAAGCAAAUACAACGAUAUUCAUAAUAUCAUUCUUUAAACUCGAAAACGCGAGACAAACUAGAAAGCUUUAAUUGAUUUCGUUAAAGAAUCAAUUAGGCUAUACAACCA--- .....................(((((.....))))).((((.....((((......)))).......))))(((((.(((((((((.......)))))))))))))).........--- ( -18.90, z-score = -2.84, R) >consensus CCAUCCCUAAGCAAAUACAACGAUAUUCAUAAUAUCAUUCUUUAAACUCGAAAACGCGAGACAAACUAGAAAGCUUUAAUUGAUUUCAUUAGAGAAUCAAUUAGGCUAUACAACCA___ .....((.((((.........(((((.....)))))..........((((......))))............))))((((((((((.......)))))))))))).............. ( -9.75 = -11.42 + 1.67)

| Location | 2,375,933 – 2,376,040 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 78.24 |
| Shannon entropy | 0.35103 |
| G+C content | 0.35122 |
| Mean single sequence MFE | -26.03 |
| Consensus MFE | -15.53 |
| Energy contribution | -18.29 |
| Covariance contribution | 2.75 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.18 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.77 |
| SVM RNA-class probability | 0.995171 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
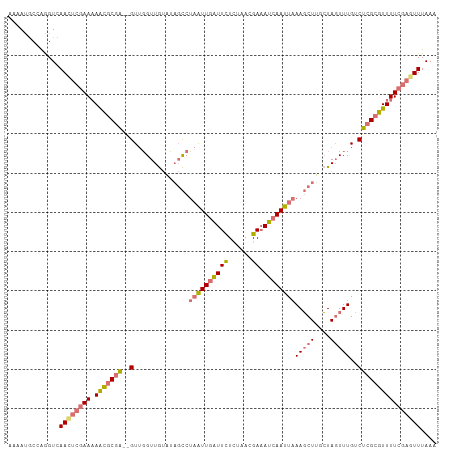
>dm3.chr2L 2375933 107 + 23011544 UUUAAACUCGAAAAAGCGAGACAAACUAGAAAGCUUUAAUUGAUUUCGUUACAGAAUCAAUUAGGCUAUACAACCAAC--UCGUCUUUUUCGAGUUGACCUGGCAUUUU ....((((((((((((((((...........(((((.(((((((((.......))))))))))))))..........)--))).))))))))))))............. ( -31.30, z-score = -5.02, R) >droAna3.scaffold_12916 2101621 106 - 16180835 AUUAAACAAGAAAACGCGAG-CAAA-UACCAAGAUUUAAUUGAUAUCAU-AGAAAACAAACGUGAAUCUGUAACAUACAACGGCGCUAUUUAGUUUGACUCGGUAUAUU .............((.((((-((((-(....(((((((.(((.......-......)))...)))))))((..(.......)..))......))))).))))))..... ( -12.72, z-score = 0.60, R) >droYak2.chr2L 2358489 107 + 22324452 UUUAAACUCGAAAACGCGAGACAAACUAGCAAGCUUUAAUUGAUUUCAUUAGAGAAUCAAUUAGGCGACACAACCAAC--UCGCGUUUUUCGAAUUGACCUGGCAUUUU .......(((((((((((((............((((.(((((((((.......)))))))))))))...........)--))))).)))))))................ ( -27.20, z-score = -3.75, R) >droSim1.chr2L 2332892 107 + 22036055 UUUAAACUCGAAAACGCGAGACAAACUAGAAAGCUUUAAUUGAUUUCGUUAAAGAAUCAAUUAGGCUAUACAACCAAC--UCGCGUUUUUCGAGUUGACCUGGCGUUUU ....((((((((((((((((...........(((((.(((((((((.......))))))))))))))..........)--))))).))))))))))............. ( -32.90, z-score = -4.55, R) >consensus UUUAAACUCGAAAACGCGAGACAAACUAGAAAGCUUUAAUUGAUUUCAUUAGAGAAUCAAUUAGGCUAUACAACCAAC__UCGCGUUUUUCGAGUUGACCUGGCAUUUU ....(((((((((((((((...............((((((((((((.......))))))))))))...............))))).))))))))))............. (-15.53 = -18.29 + 2.75)

| Location | 2,375,933 – 2,376,040 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 78.24 |
| Shannon entropy | 0.35103 |
| G+C content | 0.35122 |
| Mean single sequence MFE | -26.56 |
| Consensus MFE | -15.51 |
| Energy contribution | -16.82 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.958369 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
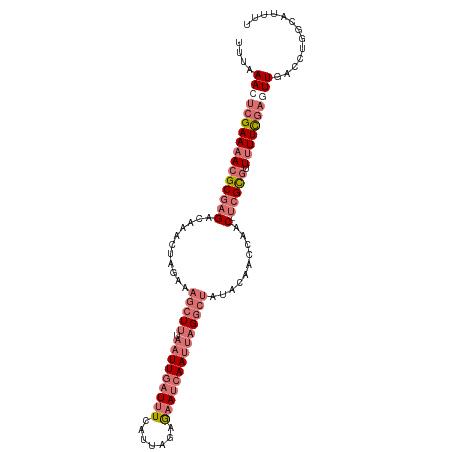
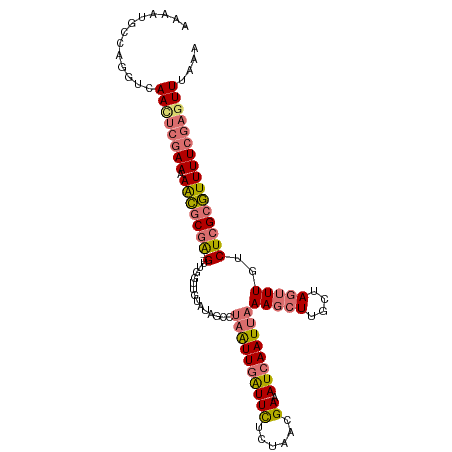
>dm3.chr2L 2375933 107 - 23011544 AAAAUGCCAGGUCAACUCGAAAAAGACGA--GUUGGUUGUAUAGCCUAAUUGAUUCUGUAACGAAAUCAAUUAAAGCUUUCUAGUUUGUCUCGCUUUUUCGAGUUUAAA .............((((((((((((.(((--(..(((......)))((((((((((......).)))))))))(((((....)))))..)))))))))))))))).... ( -29.90, z-score = -3.60, R) >droAna3.scaffold_12916 2101621 106 + 16180835 AAUAUACCGAGUCAAACUAAAUAGCGCCGUUGUAUGUUACAGAUUCACGUUUGUUUUCU-AUGAUAUCAAUUAAAUCUUGGUA-UUUG-CUCGCGUUUUCUUGUUUAAU .....(((((((.(((((((........((((.((((((.(((..((....))...)))-.))))))))))......))))).-)).)-)))).))............. ( -14.84, z-score = 0.50, R) >droYak2.chr2L 2358489 107 - 22324452 AAAAUGCCAGGUCAAUUCGAAAAACGCGA--GUUGGUUGUGUCGCCUAAUUGAUUCUCUAAUGAAAUCAAUUAAAGCUUGCUAGUUUGUCUCGCGUUUUCGAGUUUAAA .............((((((((((.(((((--(..(((......)))(((((((((.((....)))))))))))(((((....)))))..)))))))))))))))).... ( -30.00, z-score = -3.34, R) >droSim1.chr2L 2332892 107 - 22036055 AAAACGCCAGGUCAACUCGAAAAACGCGA--GUUGGUUGUAUAGCCUAAUUGAUUCUUUAACGAAAUCAAUUAAAGCUUUCUAGUUUGUCUCGCGUUUUCGAGUUUAAA .............((((((((((.(((((--(..(((......)))((((((((((......).)))))))))(((((....)))))..)))))))))))))))).... ( -31.50, z-score = -3.75, R) >consensus AAAAUGCCAGGUCAACUCGAAAAACGCGA__GUUGGUUGUAUAGCCUAAUUGAUUCUCUAACGAAAUCAAUUAAAGCUUGCUAGUUUGUCUCGCGUUUUCGAGUUUAAA .............((((((((.(((((((.................((((((((((......)).))))))))(((((....)))))...))))))))))))))).... (-15.51 = -16.82 + 1.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:11:05 2011