
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 4,674,813 – 4,674,922 |
| Length | 109 |
| Max. P | 0.728317 |

| Location | 4,674,813 – 4,674,922 |
|---|---|
| Length | 109 |
| Sequences | 7 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 69.63 |
| Shannon entropy | 0.56855 |
| G+C content | 0.59336 |
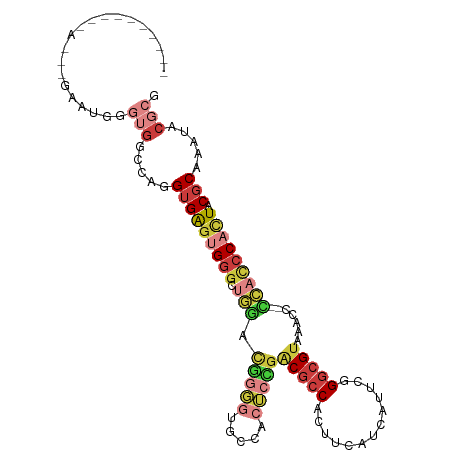
| Mean single sequence MFE | -36.45 |
| Consensus MFE | -16.59 |
| Energy contribution | -18.39 |
| Covariance contribution | 1.80 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.728317 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
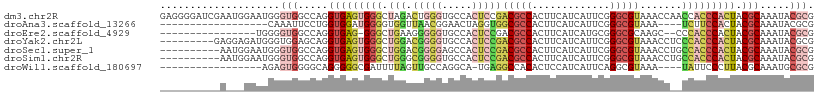
>dm3.chr2R 4674813 109 - 21146708 GAGGGGAUCGAAUGGAAUGGGUGGCCAGGUGAGUGGGCUAGACUGGGUGCCACUCCGACGCCACUUCAUCAUUCGGGCGUAAACCAACCACCCACUACGCAAAUACGCG .(((((..(((((((...(((((((...(.(((((((((......))).)))))))...)))))))..)))))))((......)).....))).)).(((......))) ( -37.70, z-score = -0.63, R) >droAna3.scaffold_13266 18538948 87 + 19884421 ------------------CAAAUUCCUGGUGGAUGGGGUGGUUAACGGAACUAGGUGGCGCCACUUCAUCAUUCGGGCGUAAA----UCUUCCACUACGCAAAUACGCG ------------------.......((((..(((((((((((..((........))...)))))))))))..))))(((((..----........)))))......... ( -25.20, z-score = -1.23, R) >droEre2.scaffold_4929 8667075 90 + 26641161 ----------------UGGGGUGGCCAGGUGAG-GGGCUGAAGGGGGUGCCACUCCGACGCCACUUCAUCAUGCGGGCGCAAGC--CCCACCCACUACGCAAAUACGCG ----------------..(((((((..((((((-((((.(..((((......))))..)))).)))))))..))((((....))--)))))))....(((......))) ( -44.90, z-score = -2.62, R) >droYak2.chr2L 17332603 100 - 22324452 ---------GAGGAGAUGGGUGGAGCAGGUGAGUGGGCUGGACGGGGUGCCACUCCGACGCCACUUCAUCAUUCGGGCGUAAACCUCCCACCCACUACGCAAAUACGCG ---------....((.(((((((.(.(((((((((((((......))).))))))..(((((.............)))))..)))))))))))))).(((......))) ( -38.02, z-score = -0.82, R) >droSec1.super_1 2312865 99 - 14215200 ----------AAUGGAAUGGGUGGCCAGGUGAGUGGGCUGGACGGGGAGCCACUCCGACGCCACUUCAUCAUUCGGGCGUAAACCUGCCACCCACUACGCAAAUACGCG ----------.......((((((((.((((((((((.((........))))))))..(((((.............)))))..))))))))))))...(((......))) ( -40.42, z-score = -1.90, R) >droSim1.chr2R 3356462 99 - 19596830 ----------AAUGGAAUGGGUGGCCAGGUGAGUGGGCUGGGCGGGGUGCCACUCCGACGCCACUUCAUCAUUCGGGCGUAAACCUGCCACCCACUACGCAAAUACGCG ----------.......((((((((.(((((((((((((......))).))))))..(((((.............)))))..))))))))))))...(((......))) ( -40.82, z-score = -1.24, R) >droWil1.scaffold_180697 1327211 87 + 4168966 -----------------AGAGUGGGGCAGGGGGCGAUUUUAGUUGCCAGGCA-UGAGGCCACACUCCAUCAUUCAGGCGUAAA----UAUUCCCUUACGCAAAUGCGCG -----------------.((((((((.((..((((((....)))))).(((.-....)))...)))).))))))..((((((.----.......))))))......... ( -28.10, z-score = -1.09, R) >consensus __________A___GAAUGGGUGGCCAGGUGAGUGGGCUGGACGGGGUGCCACUCCGACGCCACUUCAUCAUUCGGGCGUAAACC__CCACCCACUACGCAAAUACGCG ....................(((.....(((((((((.(((.(((((.....)))))(((((.............))))).......))))))))).))).....))). (-16.59 = -18.39 + 1.80)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:08:52 2011