
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 4,654,073 – 4,654,176 |
| Length | 103 |
| Max. P | 0.906049 |

| Location | 4,654,073 – 4,654,176 |
|---|---|
| Length | 103 |
| Sequences | 11 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.43 |
| Shannon entropy | 0.35991 |
| G+C content | 0.38203 |
| Mean single sequence MFE | -21.90 |
| Consensus MFE | -14.32 |
| Energy contribution | -13.77 |
| Covariance contribution | -0.54 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.906049 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 4654073 103 - 21146708 -----------CAGGUGCCGCAGAAUAUUUUAUGAAUCAUUUAUAACUCAAUAAUCCC-GUUGAGUUGAUUUCAACCAAUAAUUGAUUAUUGUUUCCGCCGUCCAGC-UGCCUUGC---- -----------((((.((.((........((((((.....))))))..((((((((..-((((.((((....))))))))....)))))))).............))-.)))))).---- ( -21.30, z-score = -1.86, R) >droSim1.chr2R_random 1593002 103 - 2996586 -----------CAGGUGCCGCAGAAUAUUUUAUGAAUCAUUUAUAACUCAAUAAUCCC-GUUGAGUUGAUUUCAACCAAUAAUUGAUUAUUGUUUCCGCCGUCCAGC-UGCCUUGC---- -----------((((.((.((........((((((.....))))))..((((((((..-((((.((((....))))))))....)))))))).............))-.)))))).---- ( -21.30, z-score = -1.86, R) >droSec1.super_1 2298299 103 - 14215200 -----------CAGGUGCCGCAGCAUAUUUUAUGAAUCAUUUAUAACUCAAUAAUCCC-GUUGAGUUGAUUUCAACCAAUAAUUGAUUAUUGUUUCCGCCGACCAGC-UGCCUUGC---- -----------((((....(((((.....((((((.....))))))..((((((((..-((((.((((....))))))))....)))))))).............))-))))))).---- ( -22.80, z-score = -2.16, R) >droYak2.chr2L 17317439 103 - 22324452 -----------CAGGUGCCGCAGAGUAUUUUAUGAAUCAUUUAUAACUCAAUAAUCCC-GUUGAGUUGAUUUCAACCAAUAAUUGAUUAUUGUUUCCGCCGUCCAGC-UGCCUUGC---- -----------((((.((.((.((((....(((((.....)))))))))(((((((..-((((.((((....))))))))....)))))))..............))-.)))))).---- ( -21.90, z-score = -1.91, R) >droEre2.scaffold_4929 8652682 103 + 26641161 -----------CAGGUGCCGCAGAAUAUUUUAUGAAUCAUUUAUAACUCAAUAAUCCC-GUUGAGUUGAUUUCGACCAAUAAUUGAUUAUUGUUUCCGCCGUCCAGC-UGCCUUGC---- -----------((((.((.((........((((((.....))))))..((((((((..-((((.((((....))))))))....)))))))).............))-.)))))).---- ( -21.00, z-score = -1.48, R) >droAna3.scaffold_13266 18518066 102 + 19884421 -----------CAGGUGCCGCAGAAUAUUUUAUGAAUCAUUUAUAACCCAAUAAUCCU-GUUGAGUUGAUUUUAACCAAUAAUUGAUUAUUGUUUCCGUCGCACGCUACCCUCC------ -----------.((((((.((.(((....((((((.....))))))..((((((((.(-((((.((((....)))))))))...)))))))).))).)).)))).)).......------ ( -21.30, z-score = -2.90, R) >dp4.chr3 13803842 107 - 19779522 ------------AGGUGCCGCAGGACAUUUUAUGAAUCAUUUAUAACUCAAUAAUCCC-GUUGAGUUGAUUUUAACCAAUAAUUGAUUAUUGUUUCCGUUUCCUCCCGUGGCCUCCCUGC ------------.((.(((((.(((..................(((((((((......-)))))))))........((((((....))))))...........))).)))))..)).... ( -24.90, z-score = -2.63, R) >droPer1.super_4 5646566 100 + 7162766 ------------AGGUGCCGCAGGACA-UUUAUGAAUCAUUUAUAACUCAAUAAUCCCGGUUGAGUUGA-UUUAAGCAAUAG-UGAUUA-UGUUUCCGUUUCCUCCCGUGCCCUGC---- ------------.......(((((...-....(((((((......(((((((.......))))))))))-)))).(((..((-.((..(-((....))).))))....))))))))---- ( -20.50, z-score = -0.69, R) >droWil1.scaffold_180697 1295671 100 + 4168966 -----------CAGGUGCCACAGAAUAUUUUAUGAAUCAUUUAUAACUCAAUAAUCCU-GUUGAGUUGAUUUUAACCAAUAAUUGAUUAUUGUUUCCGCCAACUGUCCCUAU-------- -----------..((((..(((((((...((((..........((((((((((....)-)))))))))..........))))...))).))))...))))............-------- ( -17.55, z-score = -1.75, R) >droMoj3.scaffold_6496 12969868 114 + 26866924 CCGCCUGCUAACAGGUGCCGCAGCAAAUUUUAUGAAUCAUUUAUAACUCAAUAAUCCC-GUUGAGUUGAUUUUAACCAAUAAUUGAUUAUUGUUUCCGUCGCUCAUUGUUGUUGC----- .((((((....))))))..(((((((.....((((.........((((((((......-))))))))(((...(((.(((((....))))))))...)))..))))..)))))))----- ( -28.00, z-score = -3.19, R) >droGri2.scaffold_15112 3712268 103 - 5172618 ACGUAGGCAAACAGGUGCCGUAGAAUAUUUUAUGAAUCAUUUAUAACUCAAUAAUCCU-GUUGAGUUGAUUUUAACCAAUAAUUGAUUAUUGUUUCCGUCGCCC---------------- .....(((.....(((..((((((....)))))).........((((((((((....)-)))))))))......)))((((((.....))))))......))).---------------- ( -20.40, z-score = -1.56, R) >consensus ___________CAGGUGCCGCAGAAUAUUUUAUGAAUCAUUUAUAACUCAAUAAUCCC_GUUGAGUUGAUUUUAACCAAUAAUUGAUUAUUGUUUCCGCCGCCCAGC_UGCCUUGC____ .............((((............((((((.....))))))..((((((((...((((.((((....))))))))....))))))))....)))).................... (-14.32 = -13.77 + -0.54)

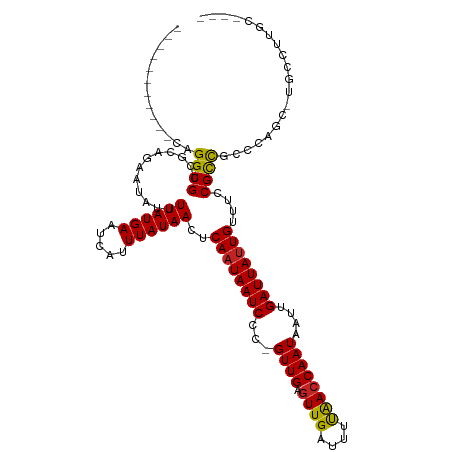
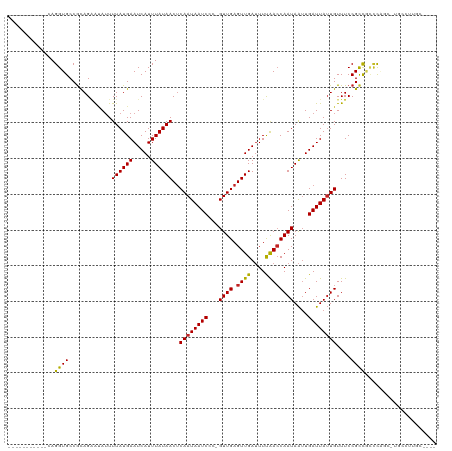
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:08:49 2011