| Sequence ID | dm3.chr2R |
|---|---|
| Location | 4,640,746 – 4,640,840 |
| Length | 94 |
| Max. P | 0.779267 |

| Location | 4,640,746 – 4,640,840 |
|---|---|
| Length | 94 |
| Sequences | 13 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 72.51 |
| Shannon entropy | 0.54895 |
| G+C content | 0.60214 |
| Mean single sequence MFE | -37.82 |
| Consensus MFE | -20.82 |
| Energy contribution | -20.32 |
| Covariance contribution | -0.51 |
| Combinations/Pair | 2.09 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.779267 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
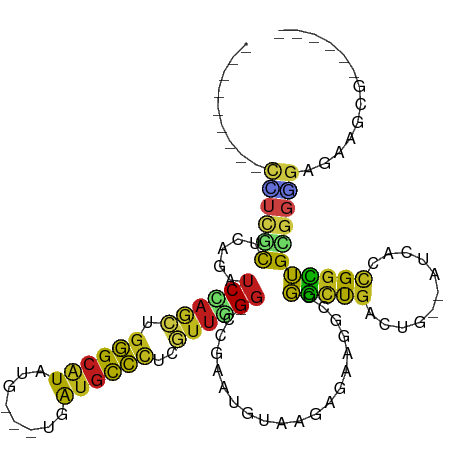
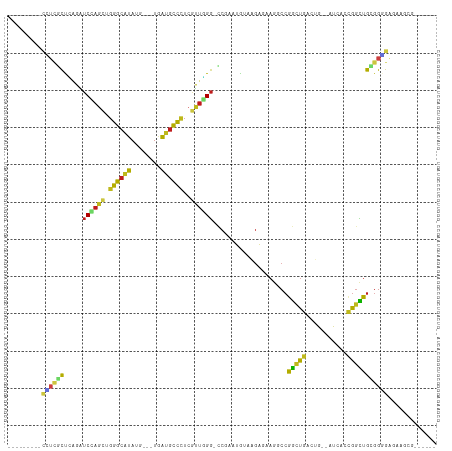
>dm3.chr2R 4640746 94 - 21146708 ---------CCUCGCCCAGAUCCAGCUGGGCAUAUG---UGAUGCCCUCGUUGGG-CCGAAUGUAAGAAAAGGCCGGCUGGCUG--AUCGCCGGCUGCGGGGAGAAGCG------ ---------.(((.(((.(..(((((.((((((...---..))))))..))))).-.)..............((.(((((((..--...))))))))))))))).....------ ( -42.10, z-score = -1.69, R) >droSim1.chr2R 3328562 94 - 19596830 ---------CCUCGCCCAGAUCCAGCUGGGCAUAUG---UGAUGCCCUCGUUGGG-CCUAAUGUAAGAGAAUGCCGGCUGACUG--AUCACCGGCUGCGGGGAGAAGCG------ ---------((((((..((..(((((.((((((...---..))))))..))))).-.)).............(((((.(((...--.)))))))).)))))).......------ ( -40.20, z-score = -2.31, R) >droSec1.super_1 2285053 94 - 14215200 ---------CCUCGCCCAGAUCCAGCUGGGCAUAUG---UGAUGCCUUCGUUGGG-CCUAAUGUAAGAGAAGGCCGGUUGGCUG--AUUACCGGCUGCGGGGAGAAGCG------ ---------((((((..((..(((((.((((((...---..))))))..))))).-.))............(((((((......--...))))))))))))).......------ ( -38.80, z-score = -1.61, R) >droYak2.chr2L 17304332 94 - 22324452 ---------CCUCUCCCAGGUCCAGCUGGGCAUAUG---UGAUGCCCUCGUUGGG-GCGAAUGUAAGAGAAGGCCGGCUGGCUG--AUCGCCGGCUGCGGGGAGAAGCG------ ---------..((((((.(.((((((.((((((...---..))))))..))))))-.)..............((.(((((((..--...))))))))).))))))....------ ( -44.60, z-score = -1.92, R) >droEre2.scaffold_4929 8639536 94 + 26641161 ---------CCUCGCCCAGAUCCAGCUGGGCAUAUG---UGAUGCCCUCGUUGGG-CCGAAUGUAAGAGAAGGCCGGCUGGCUG--AUCACCGGCUGCGGGGAGAAGCG------ ---------((((((...(..(((((.((((((...---..))))))..))))).-.).............((((((.(((...--.))))))))))))))).......------ ( -40.00, z-score = -1.18, R) >droAna3.scaffold_13266 18504010 94 + 19884421 ---------CUUCACCCAAAUCCAACUGGGCGUAGG---UAAUGCCGUCGUUGGG-CCGUAUGUAGGAGAAGCCCGGUUGACUA--AUCACCGGCUGUGGGGAAAAUCG------ ---------.(((.((((..((((((..(((((...---..)))))...))))))-((.......))...((((.(((.((...--.))))))))).))))))).....------ ( -29.30, z-score = -0.11, R) >dp4.chr3 13789787 103 - 19779522 UCUCGUCCAGCGGACUGUGGUCCAGCUGGGCGUAUG---UGAUGCCCUCGUUGGG-CCGGAUGUAGGAGAAGCCGGGUUGGCUG--AUGUAGGGCUGUGGAGAGAAGCG------ (((((((((((((((....)))).))))))).....---....((((((((..(.-((((.....((.....))...)))))..--))).)))))......))))....------ ( -44.30, z-score = -2.14, R) >droPer1.super_4 5632372 103 + 7162766 UCUCGUCCAGCGGACUGUGGUCCAGCUGGGCGUAUG---UGAUGCCCUCAUUGGG-CCGGAUGUAGGAGAAGCCCGGCUGGCUG--AUGUACGGCUGCGGAGAGAAGCG------ ((((.(((.((((.((((((.((((((((((.((((---(...((((.....)))-)...)))))......)))))))))))).--....)))))))))))))))....------ ( -46.30, z-score = -1.89, R) >droWil1.scaffold_180697 1276307 94 + 4168966 ---------CAUUGCUUAAGUCGAGUUGGGCAUAGG---UGAUGCCCUCAUUGGG-CCUUAUAUACGAAAAGGCCGGCUGACUA--AUUACUGGCUGUGGUGAGAAACG------ ---------(((..(...((((.(((.((((((...---..))))))(((.(.((-((((.........)))))).).)))...--...))))))))..))).......------ ( -29.10, z-score = -1.48, R) >droVir3.scaffold_13324 2842580 94 - 2960039 ---------CGUCGCUGAGAUCGAGCUGGGCGUAGG---UGAUGCCCUCGUUUGG-CCGUAUAUAGGAAAAAGCCGGCUGACUA--AUGACCGGUUGGGGUGAGAAGCG------ ---------..(((((..(..(((((.((((((...---..))))))..))))).-.).............(((((((......--..).))))))..)))))......------ ( -28.90, z-score = 0.03, R) >droMoj3.scaffold_6496 12949489 94 + 26866924 ---------CAUCGCUCAGAUCAAGCUGGGCGUAGG---UGAUGCCCUCGUUGGG-CCGUAUAUAGGAGAAGGCCGGCUGACUA--AUGACCGGCUGUGGUGAGAAACG------ ---------((((((((((......))))))....(---((..((((.....)))-)..))).........(((((((......--..).))))))..)))).......------ ( -30.50, z-score = -0.19, R) >droGri2.scaffold_15112 3695801 97 - 5172618 ---------CGUCGCUGUGAUCGAGCUGGGCAUAGGUGAUGAUGCCCUCAUUGGG-UCGUAUAUACGAGAAGCCCGGCUGACUA--AUGACCGGCUGUGGCGAGAAGCG------ ---------..((((..((..((((((((((.............(((.....)))-((((....))))...))))))))..(..--..)..))..))..))))......------ ( -32.60, z-score = -0.46, R) >anoGam1.chr3L 3551836 109 + 41284009 ------UCGUCUCGUUGGCGACGACCUGCGCGUACACGAUCGGCUGCUCGCCCGAUCCGCGGGUGUACGAAAACCCACCGCCCACGAUCUGCGAUGGCGGAGGGGAGGUGUGCCG ------(((((.....)))))..(((((((.(....)(((((((.....).)))))))))))))(((((....(((.(((((..((.....))..))))).)))....))))).. ( -44.90, z-score = 0.26, R) >consensus _________CCUCGCUCAGAUCCAGCUGGGCAUAUG___UGAUGCCCUCGUUGGG_CCGAAUGUAAGAGAAGGCCGGCUGACUG__AUCACCGGCUGCGGGGAGAAGCG______ .........((((((......(((((.((((((........))))))..))))).....................(((((...........)))))))))))............. (-20.82 = -20.32 + -0.51)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:08:46 2011