| Sequence ID | dm3.chr2L |
|---|---|
| Location | 2,364,914 – 2,365,018 |
| Length | 104 |
| Max. P | 0.802770 |

| Location | 2,364,914 – 2,365,018 |
|---|---|
| Length | 104 |
| Sequences | 7 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 62.33 |
| Shannon entropy | 0.74381 |
| G+C content | 0.42818 |
| Mean single sequence MFE | -23.03 |
| Consensus MFE | -8.76 |
| Energy contribution | -8.81 |
| Covariance contribution | 0.05 |
| Combinations/Pair | 1.67 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.802770 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
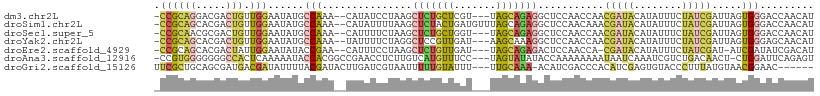
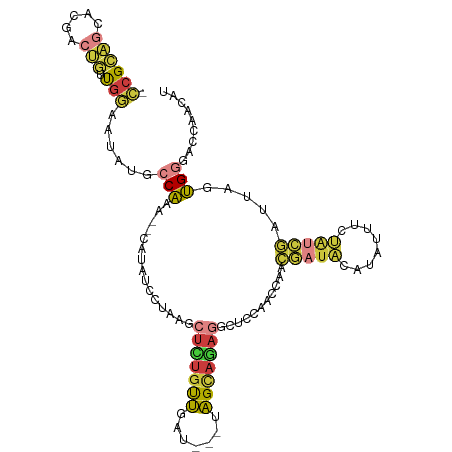
>dm3.chr2L 2364914 104 + 23011544 -CCGCAGGACGACUGUUGGAAUAUGCCAAA--CAUAUCCUAAGCUCUGCUCGU---UAGCAGAGGCUCCAACCAACGAUACAUAUUUCUAUCGAUUAGUGGGACCAACAU -((((.........(((((((((((.....--)))))......(((((((...---.)))))))..))))))...(((((........)))))....))))......... ( -27.80, z-score = -2.09, R) >droSim1.chr2L 2321750 107 + 22036055 -CCGCAGCACGACUGUUGGAAUAUGCCAAA--CAUAUUUUAAGCUCUACUGAUGUUUAGCAGAGGCUCCAACAAACGAUACAUAUUUCUAUCGAUUAGUGGGACCAACAU -((((........((((((((((((.....--)))))......((((.((((...)))).))))..)))))))..(((((........)))))....))))......... ( -24.60, z-score = -1.75, R) >droSec1.super_5 536515 104 + 5866729 -CCGCAACGCGACUGUUGGAAUAUGCCAAA--CAUUUUCUAAGCUCUGCUGGU---UAGCAGAGGCUCCAACCAACGAUACAUAUUUCUAUCGAUUAGUGGGACCAACAU -.(((...)))..((((((...........--...........(((((((...---.)))))))..((((.....(((((........))))).....)))).)))))). ( -26.60, z-score = -1.68, R) >droYak2.chr2L 2347352 104 + 22324452 -CCGCAGCACGACUGUUGGAAUAUGCCAAA--UAUUUUCUAGGCUCCGUUGAU---AAGCAAAGGCUCCAACCAACGAUACAUAUUUCUAUCGAUUAGUGGGAGCAACAU -...(((((....)))))((((((.....)--))))).....(((((((((..---.(((....))).))))...(((((........))))).......)))))..... ( -22.50, z-score = -0.45, R) >droEre2.scaffold_4929 2404226 102 + 26641161 -CCGCAGCACGACUAUUGGAAUAUACCGAA--CAUUUCCUAAGCUCUGUUGAU---UAGCAGAGACUCCAACCA-CGAUACAUAUUUCUAUCGAU-AUCGAUAUCGACAU -........(((.((((((...........--.........((((((((....---..)))))).)).......-(((((........)))))..-.))))))))).... ( -17.70, z-score = -1.07, R) >droAna3.scaffold_12916 10089140 105 - 16180835 -CCGUGGGGGGGCCACUCAAAAAUACCACGGCCGAACCUCUUGUCAUGUUUCC---UAGUAUAUACCAAAAAAAAUAAUCAAAUCGUCUGACAACU-CUGGAUUCAGAGU -....((((.((((...............))))...)))).(((((.......---................................)))))(((-(((....)))))) ( -18.99, z-score = 0.88, R) >droGri2.scaffold_15126 2341266 100 - 8399593 UUCGCUGCAGCGAUGACGAUAUUUUACGAUACUUGAUCGUAAUUUUUGUAUUU---UUGCAAA-ACAUCGACCCACAUCGAGUGUACCCUUUAUGUAACGGAAC------ ((((.((((.(((((........(((((((.....))))))).(((((((...---.))))))-))))))...(((.....))).........)))).))))..------ ( -23.00, z-score = -2.12, R) >consensus _CCGCAGCACGACUGUUGGAAUAUGCCAAA__CAUAUCCUAAGCUCUGUUGAU___UAGCAGAGGCUCCAACCAACGAUACAUAUUUCUAUCGAUUAGUGGGACCAACAU .((((((.....))).)))......(((...............(((((((.......)))))))...........(((((........))))).....)))......... ( -8.76 = -8.81 + 0.05)

| Location | 2,364,914 – 2,365,018 |
|---|---|
| Length | 104 |
| Sequences | 7 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 62.33 |
| Shannon entropy | 0.74381 |
| G+C content | 0.42818 |
| Mean single sequence MFE | -27.30 |
| Consensus MFE | -9.04 |
| Energy contribution | -6.85 |
| Covariance contribution | -2.19 |
| Combinations/Pair | 2.09 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.585923 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
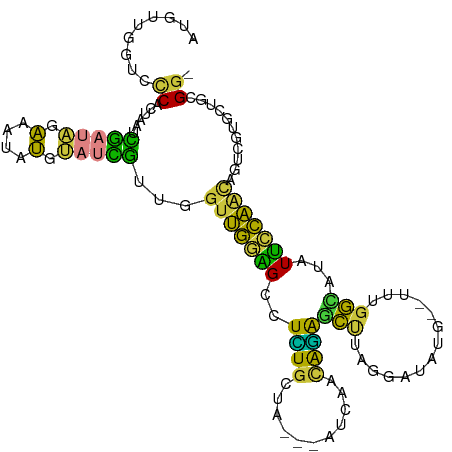
>dm3.chr2L 2364914 104 - 23011544 AUGUUGGUCCCACUAAUCGAUAGAAAUAUGUAUCGUUGGUUGGAGCCUCUGCUA---ACGAGCAGAGCUUAGGAUAUG--UUUGGCAUAUUCCAACAGUCGUCCUGCGG- .(((((.(((.(((((.(((((.(....).)))))))))).)))((.((((((.---...))))))))...(((((((--.....))))))))))))..((.....)).- ( -33.10, z-score = -2.17, R) >droSim1.chr2L 2321750 107 - 22036055 AUGUUGGUCCCACUAAUCGAUAGAAAUAUGUAUCGUUUGUUGGAGCCUCUGCUAAACAUCAGUAGAGCUUAAAAUAUG--UUUGGCAUAUUCCAACAGUCGUGCUGCGG- .((((((..(((.(((.(((((.(....).))))).))).)))(((.((((((.......)))))))))...((((((--.....))))))))))))..((.....)).- ( -26.90, z-score = -1.28, R) >droSec1.super_5 536515 104 - 5866729 AUGUUGGUCCCACUAAUCGAUAGAAAUAUGUAUCGUUGGUUGGAGCCUCUGCUA---ACCAGCAGAGCUUAGAAAAUG--UUUGGCAUAUUCCAACAGUCGCGUUGCGG- .(((((((((.(((((.(((((.(....).)))))))))).)))(((((((((.---...))))))((.........)--)..))).....))))))..(((...))).- ( -31.20, z-score = -1.94, R) >droYak2.chr2L 2347352 104 - 22324452 AUGUUGCUCCCACUAAUCGAUAGAAAUAUGUAUCGUUGGUUGGAGCCUUUGCUU---AUCAACGGAGCCUAGAAAAUA--UUUGGCAUAUUCCAACAGUCGUGCUGCGG- .....(((((.(((((.(((((.(....).)))))))))).)))))........---.....((.(((...((.....--.((((......))))...))..))).)).- ( -26.90, z-score = -1.03, R) >droEre2.scaffold_4929 2404226 102 - 26641161 AUGUCGAUAUCGAU-AUCGAUAGAAAUAUGUAUCG-UGGUUGGAGUCUCUGCUA---AUCAACAGAGCUUAGGAAAUG--UUCGGUAUAUUCCAAUAGUCGUGCUGCGG- .((((((((....)-)))))))...........((-..((..(((.(((((...---.....)))))))).((((.((--(.....))))))).........))..)).- ( -24.40, z-score = -0.33, R) >droAna3.scaffold_12916 10089140 105 + 16180835 ACUCUGAAUCCAG-AGUUGUCAGACGAUUUGAUUAUUUUUUUUGGUAUAUACUA---GGAAACAUGACAAGAGGUUCGGCCGUGGUAUUUUUGAGUGGCCCCCCCACGG- ((((((....)))-))).(((..((..((((.((((.(((((((((....))))---))))).))))))))..))..)))(((((..................))))).- ( -25.57, z-score = -0.37, R) >droGri2.scaffold_15126 2341266 100 + 8399593 ------GUUCCGUUACAUAAAGGGUACACUCGAUGUGGGUCGAUGU-UUUGCAA---AAAUACAAAAAUUACGAUCAAGUAUCGUAAAAUAUCGUCAUCGCUGCAGCGAA ------......((((((...(((....))).))))))(.((((((-(((((..---..((((...............)))).))))))))))).).((((....)))). ( -23.06, z-score = -1.43, R) >consensus AUGUUGGUCCCACUAAUCGAUAGAAAUAUGUAUCGUUGGUUGGAGCCUCUGCUA___AUCAACAGAGCUUAGGAUAUG__UUUGGCAUAUUCCAACAGUCGUGCUGCGG_ .........((......(((((.(....).)))))...(((((((..((((...........))))(((..............)))...)))))))...........)). ( -9.04 = -6.85 + -2.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:11:03 2011