| Sequence ID | dm3.chr2R |
|---|---|
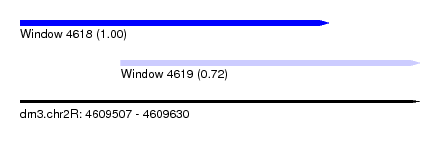
| Location | 4,609,507 – 4,609,630 |
| Length | 123 |
| Max. P | 0.996762 |

| Location | 4,609,507 – 4,609,602 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 85.82 |
| Shannon entropy | 0.25279 |
| G+C content | 0.48743 |
| Mean single sequence MFE | -36.92 |
| Consensus MFE | -25.22 |
| Energy contribution | -25.92 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.61 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.98 |
| SVM RNA-class probability | 0.996762 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
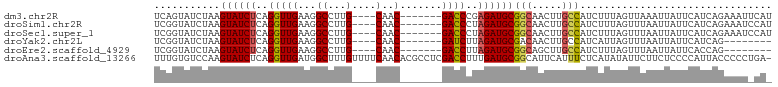
>dm3.chr2R 4609507 95 + 21146708 UUGCUCAGAUGGCUGCCGUUGGAUAUCCGAUUCAGUAUCUAAGUAUCUCAGGUUGAAGGCCUUG----CAAC-------GACCCGAGAUGCGGCA---ACUUGCCAUCU ......(((((((((((.((((((((........))))))))(((((((.(((((...((...)----)..)-------)))).)))))))))))---....))))))) ( -39.30, z-score = -4.19, R) >droSim1.chr2R 3293306 95 + 19596830 UUGCUCAGAUGGCUGCCGUUGAAUACCCGAUUCGGUAUCUAAGUAUCUCAGGUUGAAGGCCUUG----CAAC-------GACCCUAGAUGCGGCA---ACUUGCCAUCU ......(((((((((((.(((.(((((......))))).)))((((((..(((((...((...)----)..)-------))))..))))))))))---....))))))) ( -34.50, z-score = -3.16, R) >droSec1.super_1 2253933 95 + 14215200 UUGCUCAGAUGGCUGCCGUUGGAUAUCCGAUUCGGUAUCUAAGUAUCUCAGGUUGAAGGCCUUG----CAAC-------GACCCUAGAUGCGGCA---ACUUGCCAUCU ......(((((((((((.(((((((((......)))))))))((((((..(((((...((...)----)..)-------))))..))))))))))---....))))))) ( -38.10, z-score = -3.73, R) >droYak2.chr2L 17273156 95 + 22324452 UUGCUCAGAUGGCUGCCAUUGGAUAUCCCAUUCGGUAUCUAAGUAUCUCAGGUUGAAGGCCUUG----CAAC-------GAUCUUAGAUGCGACA---ACUUGCCAUCA .......((((((.....(((((((((......)))))))))((((((.((((((...((...)----)..)-------))))).))))))....---....)))))). ( -32.10, z-score = -2.77, R) >droEre2.scaffold_4929 8608844 95 - 26641161 UUGCUCAGAUGGCUGCCGUUGGAUAUCCGAUUCGGUAUCUAAGUAUCUCAGGUUGAAGGCCUUG----CAAC-------GACCUUAGAUGCGGCA---GCUUGCCAUCU ......(((((((((((.(((((((((......)))))))))((((((.((((((...((...)----)..)-------))))).))))))))))---....))))))) ( -40.10, z-score = -3.87, R) >droAna3.scaffold_13266 18472240 109 - 19884421 UUGCACAAAUGGCCGUGCUUGGAUAUCCGAUUUUGUGUCCAAGUAUCUCAGGUUGAUGGCUUUGUUUUCAACACGCCUCGACCUUUGAUGCGGCAUUCAUUUCUCAUAU ...........(((((((((((((((........))))))))))(((..(((((((.(((..(((.....))).))))))))))..))))))))............... ( -37.40, z-score = -3.94, R) >consensus UUGCUCAGAUGGCUGCCGUUGGAUAUCCGAUUCGGUAUCUAAGUAUCUCAGGUUGAAGGCCUUG____CAAC_______GACCCUAGAUGCGGCA___ACUUGCCAUCU .......((((((((((.(((((((((......)))))))))((((((..((((.......(((....)))........))))..)))))))))).......)))))). (-25.22 = -25.92 + 0.70)

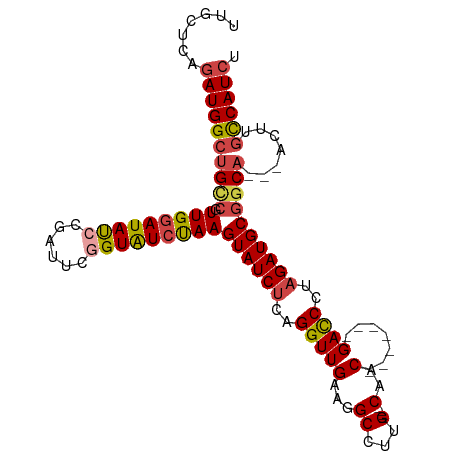
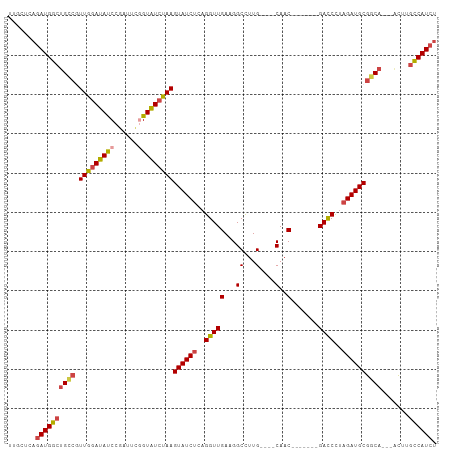
| Location | 4,609,538 – 4,609,630 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 77.33 |
| Shannon entropy | 0.40605 |
| G+C content | 0.41713 |
| Mean single sequence MFE | -22.37 |
| Consensus MFE | -10.30 |
| Energy contribution | -10.41 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.716140 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 4609538 92 + 21146708 UCAGUAUCUAAGUAUCUCAGGUUGAAGGCCUUG----CAAC-------GACCCGAGAUGCGGCAACUUGCCAUCUUUAGUUAAAUUAUUCAUCAGAAAUUCAU ....((.(((((((((((.(((((...((...)----)..)-------)))).)))))))(((.....)))....)))).))..................... ( -23.60, z-score = -2.49, R) >droSim1.chr2R 3293337 92 + 19596830 UCGGUAUCUAAGUAUCUCAGGUUGAAGGCCUUG----CAAC-------GACCCUAGAUGCGGCAACUUGCCAUCUUUAGUUUAAUUAUUCAUCAGAAAUCCAU ..((..(((..((((((..(((((...((...)----)..)-------))))..))))))(((.....)))......................)))...)).. ( -22.40, z-score = -1.83, R) >droSec1.super_1 2253964 92 + 14215200 UCGGUAUCUAAGUAUCUCAGGUUGAAGGCCUUG----CAAC-------GACCCUAGAUGCGGCAACUUGCCAUCUUUAGUUUAAUUAUUCAUCAGAAAUCCAU ..((..(((..((((((..(((((...((...)----)..)-------))))..))))))(((.....)))......................)))...)).. ( -22.40, z-score = -1.83, R) >droYak2.chr2L 17273187 84 + 22324452 UCGGUAUCUAAGUAUCUCAGGUUGAAGGCCUUG----CAAC-------GAUCUUAGAUGCGACAACUUGCCAUCAUUAGUUUAAUUAUUCAUCAG-------- ..((((.....((((((.((((((...((...)----)..)-------))))).)))))).......))))........................-------- ( -17.80, z-score = -1.34, R) >droEre2.scaffold_4929 8608875 84 - 26641161 UCGGUAUCUAAGUAUCUCAGGUUGAAGGCCUUG----CAAC-------GACCUUAGAUGCGGCAGCUUGCCAUCUUUAGUUUAAUUAUUCACCAG-------- ..(((......((((((.((((((...((...)----)..)-------))))).))))))(((.....)))...................)))..-------- ( -24.40, z-score = -2.56, R) >droAna3.scaffold_13266 18472271 102 - 19884421 UUUGUGUCCAAGUAUCUCAGGUUGAUGGCUUUGUUUUCAACACGCCUCGACCUUUGAUGCGGCAUUCAUUUCUCAUAUAUUCUUCUCCCCAUUACCCCCUGA- ...(((((...(((((..(((((((.(((..(((.....))).))))))))))..)))))))))).....................................- ( -23.60, z-score = -2.82, R) >consensus UCGGUAUCUAAGUAUCUCAGGUUGAAGGCCUUG____CAAC_______GACCCUAGAUGCGGCAACUUGCCAUCUUUAGUUUAAUUAUUCAUCAGAAAU_CA_ ...........((((((..((((.......(((....)))........))))..))))))(((.....)))................................ (-10.30 = -10.41 + 0.11)
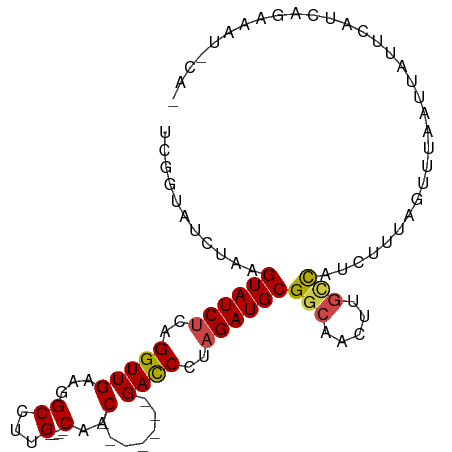
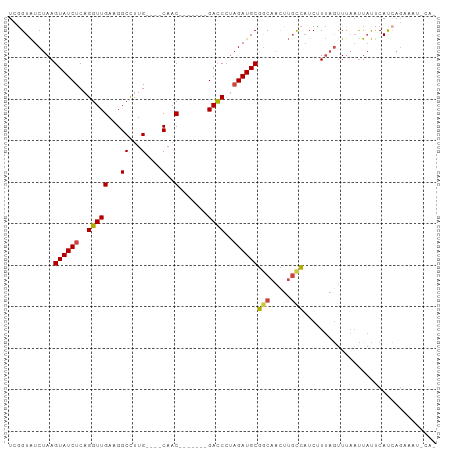
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:08:41 2011