| Sequence ID | dm3.chr2R |
|---|---|
| Location | 4,607,000 – 4,607,126 |
| Length | 126 |
| Max. P | 0.896379 |

| Location | 4,607,000 – 4,607,126 |
|---|---|
| Length | 126 |
| Sequences | 5 |
| Columns | 138 |
| Reading direction | forward |
| Mean pairwise identity | 75.84 |
| Shannon entropy | 0.41569 |
| G+C content | 0.37808 |
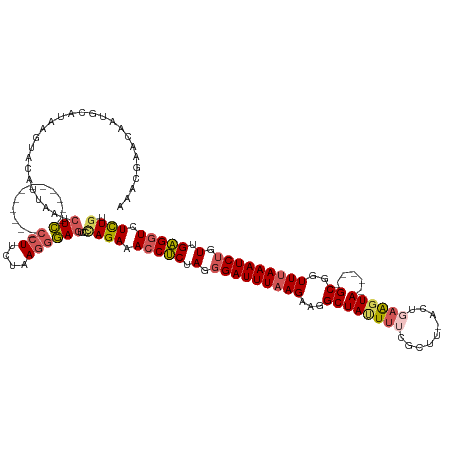
| Mean single sequence MFE | -32.78 |
| Consensus MFE | -16.33 |
| Energy contribution | -16.82 |
| Covariance contribution | 0.49 |
| Combinations/Pair | 1.16 |
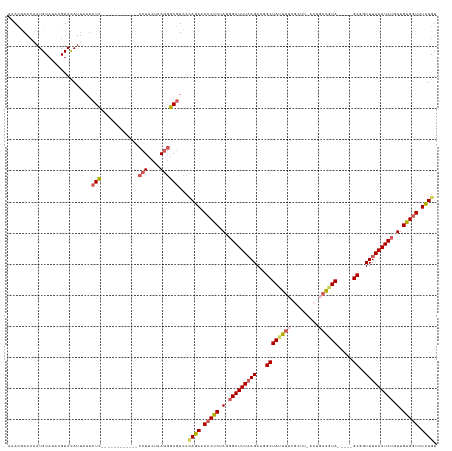
| Mean z-score | -2.13 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.846793 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
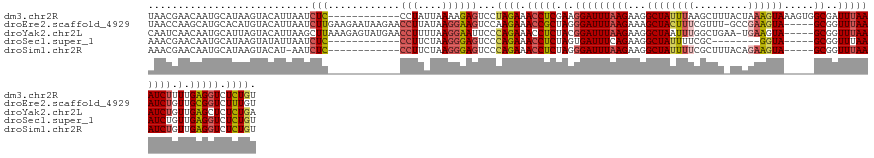
>dm3.chr2R 4607000 126 + 21146708 UAACGAACAAUGCAUAAGUACAUUAAUCUC------------CCUAUUAAAAGAGUCCUAGAAACCUCGAAGGAUUUAAGAAGGCUAUUUAAGCUUUACUAAAGUAAAGUGGCGAUUUAAAUCUUUUGAGGUCUCUGU ....((..((((........))))..))..------------................((((.(((((((((((((((((...(((......(((((((....))))))))))..))))))))))))))))).)))). ( -32.20, z-score = -2.90, R) >droEre2.scaffold_4929 8606363 132 - 26641161 UAACCAAGCAUGCACAUGUACAUUAAUCUUGAAGAAUAAGAACCUUAUAAGGAAGUCCAAGAAACCGCUAGGGAUUUAAGAAAGCUACUUUCGUUU-GCCGAAGUA-----GCGGUUUAAAUCUGUUGCGGUCUUUGU ...((..((((....)))).......(((((.....))))).........)).......(((.(((((.(.(((((((((...((((((((.(...-.).))))))-----))..))))))))).).))))).))).. ( -34.70, z-score = -2.14, R) >droYak2.chr2L 17270634 132 + 22324452 CAAUCAACAAUGCAUUAGUACAUUAAGCUUAAAGAGUAUGAACCUUUUAAGGAAUUCCCAGAAACCUCUACGGAUUUAAGAAGGCUAAUUUGGCUGAA-UGAAGUA-----GCGGUUUAAAUCUGUUGAGCUCUCUGA ...((...((((........))))...(((((((.((....)).))))))))).....((((...(((.(((((((((((...((((.((((......-)))).))-----))..))))))))))).)))...)))). ( -27.50, z-score = -0.15, R) >droSec1.super_1 2251441 113 + 14215200 AAACGAACAAUGCAUAAGUAUAUUAAUCUC------------CCUUCUAAGGGAGUCCCAGAAACCUCUAGUGAUUUCAGAAGGCUAUUUUCGC--------GGUA-----GCGGUUUAAAUCUGUUGAGGUCUCUGU .........((((....))))......(((------------(((....))))))...((((.(((((....(((((.(((..((((((.....--------))))-----))..))))))))....))))).)))). ( -31.90, z-score = -2.28, R) >droSim1.chr2R 3290807 120 + 19596830 AAACGAACAAUGCAUAAGUACAU-AAUCUC------------CCUUCUAAGGGAGUCCCAGAAACCUCUAGGGAUUUAAGAAGGCUAUUUUCGCUUUACAGAAGUA-----GCGGUUUAAAUCUGUUGAGGUCUCUGU .......................-...(((------------(((....))))))...((((.(((((.(.(((((((((...(((((((..(.....)..)))))-----))..))))))))).).))))).)))). ( -37.60, z-score = -3.20, R) >consensus AAACGAACAAUGCAUAAGUACAUUAAUCUC____________CCUUCUAAGGGAGUCCCAGAAACCUCUAGGGAUUUAAGAAGGCUAUUUUCGCUU_ACUGAAGUA_____GCGGUUUAAAUCUGUUGAGGUCUCUGU ..........................................................((((.(((((.(.(((((((((...((.......((((.....))))......))..))))))))).).))))).)))). (-16.33 = -16.82 + 0.49)
| Location | 4,607,000 – 4,607,126 |
|---|---|
| Length | 126 |
| Sequences | 5 |
| Columns | 138 |
| Reading direction | reverse |
| Mean pairwise identity | 75.84 |
| Shannon entropy | 0.41569 |
| G+C content | 0.37808 |
| Mean single sequence MFE | -32.20 |
| Consensus MFE | -14.51 |
| Energy contribution | -15.99 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.896379 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 4607000 126 - 21146708 ACAGAGACCUCAAAAGAUUUAAAUCGCCACUUUACUUUAGUAAAGCUUAAAUAGCCUUCUUAAAUCCUUCGAGGUUUCUAGGACUCUUUUAAUAGG------------GAGAUUAAUGUACUUAUGCAUUGUUCGUUA ..(((((((((.((.(((((((........(((((....)))))(((.....)))....))))))).)).)))))))))....((((......)))------------).((.(((((((....))))))).)).... ( -30.80, z-score = -2.44, R) >droEre2.scaffold_4929 8606363 132 + 26641161 ACAAAGACCGCAACAGAUUUAAACCGC-----UACUUCGGC-AAACGAAAGUAGCUUUCUUAAAUCCCUAGCGGUUUCUUGGACUUCCUUAUAAGGUUCUUAUUCUUCAAGAUUAAUGUACAUGUGCAUGCUUGGUUA ....(((((((....(((((((...((-----(((((((..-...)).)))))))....)))))))....)))))))...((((((.......)))))).......(((((....(((((....))))).)))))... ( -35.60, z-score = -3.15, R) >droYak2.chr2L 17270634 132 - 22324452 UCAGAGAGCUCAACAGAUUUAAACCGC-----UACUUCA-UUCAGCCAAAUUAGCCUUCUUAAAUCCGUAGAGGUUUCUGGGAAUUCCUUAAAAGGUUCAUACUCUUUAAGCUUAAUGUACUAAUGCAUUGUUGAUUG (((((((.(((.((.(((((((...((-----((.....-...........))))....))))))).)).))).)))))))......((((((..((....))..))))))..(((((((....)))))))....... ( -28.99, z-score = -1.42, R) >droSec1.super_1 2251441 113 - 14215200 ACAGAGACCUCAACAGAUUUAAACCGC-----UACC--------GCGAAAAUAGCCUUCUGAAAUCACUAGAGGUUUCUGGGACUCCCUUAGAAGG------------GAGAUUAAUAUACUUAUGCAUUGUUCGUUU .((((((((((....(((((.....((-----((..--------.......))))......)))))....))))))))))...((((((....)))------------)))........................... ( -30.60, z-score = -2.51, R) >droSim1.chr2R 3290807 120 - 19596830 ACAGAGACCUCAACAGAUUUAAACCGC-----UACUUCUGUAAAGCGAAAAUAGCCUUCUUAAAUCCCUAGAGGUUUCUGGGACUCCCUUAGAAGG------------GAGAUU-AUGUACUUAUGCAUUGUUCGUUU .((((((((((....(((((((...((-----((.((.((.....)).)).))))....)))))))....))))))))))(((((((((....)))------------)))...-(((((....)))))..))).... ( -35.00, z-score = -2.85, R) >consensus ACAGAGACCUCAACAGAUUUAAACCGC_____UACUUCAGU_AAGCGAAAAUAGCCUUCUUAAAUCCCUAGAGGUUUCUGGGACUCCCUUAAAAGG____________GAGAUUAAUGUACUUAUGCAUUGUUCGUUA .((((((((((....(((((((...((..........................))....)))))))....)))))))))).................................(((((((....)))))))....... (-14.51 = -15.99 + 1.48)



Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:08:40 2011