
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 4,562,340 – 4,562,401 |
| Length | 61 |
| Max. P | 0.533553 |

| Location | 4,562,340 – 4,562,401 |
|---|---|
| Length | 61 |
| Sequences | 11 |
| Columns | 74 |
| Reading direction | forward |
| Mean pairwise identity | 71.23 |
| Shannon entropy | 0.55658 |
| G+C content | 0.38284 |
| Mean single sequence MFE | -8.57 |
| Consensus MFE | -6.27 |
| Energy contribution | -6.12 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.12 |
| Mean z-score | -0.23 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.533553 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 4562340 61 + 21146708 ------------AAAAGGCAGCAAGAUGUUCAUCGAAGACGAUGCGCAGUAAGUA-AACACAUUUAUUAUAUAA ------------...............((.(((((....))))).))(((((((.-.....)))))))...... ( -8.20, z-score = -0.54, R) >droGri2.scaffold_15245 5007946 57 - 18325388 ------------AAAAGUCGGCAAGAUGUUCAUCGAAGAUGAUGCGCAGUAAGUA--CAAUUUCCAUUUAA--- ------------........((....(((.((((......)))).)))....)).--..............--- ( -6.00, z-score = 0.90, R) >droMoj3.scaffold_6496 11347462 56 - 26866924 ------------AAAGCUUGGCAACAUGUUCAUCGAAGAUGAUGCGCAGUAAGUA--CCGUUUC-AUUCAA--- ------------...(((.((((.(((.(((...))).))).))).)))).....--.......-......--- ( -8.50, z-score = 0.57, R) >droVir3.scaffold_12875 11406778 56 + 20611582 ------------GAAGCUCGGCAAGAUGUUCAUCGAAGAUGAUGCCCAGUAAGUA--CUCUCCC-AUUGAA--- ------------...(((.((((..((.(((...))).))..)))).))).....--.......-......--- ( -9.80, z-score = -0.06, R) >droWil1.scaffold_180700 1636975 57 + 6630534 ------------AAAAGCCAUCAAGAUGUUCAUCGAAGAUGAUGCGCAGUAAGUACAACCUAAUUCUUA----- ------------....(((((((.(((....))).....))))).))......................----- ( -8.00, z-score = -0.61, R) >droAna3.scaffold_13266 18434424 59 - 19884421 ----------AAAAAAACCGCCAACAUGUUCAUCGAAGAUGAUGCGCAGUAAGUAGUAUCUAUAA--GAUA--- ----------......((..(..((.(((.((((......)))).)))))..)..))........--....--- ( -8.00, z-score = -0.89, R) >dp4.chr3 13703076 74 + 19779522 AAACCAAGAGAAAGCACACGACGAGAUGUUCAUCGAAGAUGAUGCGCAGUAAGUAGUACCAAUGCCCGAUACCG .............(((.((.((....(((.((((......)))).)))....)).)).....)))......... ( -8.40, z-score = 0.34, R) >droSim1.chr2R 3223376 61 + 19596830 ------------AAAAGGCAGCAAGAUGUUCAUCGAAGACGAUGCGCAGUAAGUA-AAGAUAUCUAUUAUAUAA ------------........((....(((.(((((....))))).)))....)).-.................. ( -8.10, z-score = -0.12, R) >droSec1.super_1 2206848 61 + 14215200 ------------AAAAGGCAGCAAGAUGUUCAUCGAAGACGAUGCGCAGUAAGUA-AAGACAUCUAUUAUAUAA ------------...........((((((((((((....))))).((.....)).-..)))))))......... ( -9.90, z-score = -0.91, R) >droYak2.chr2L 17222725 61 + 22324452 ------------AAGAAGCAUCAAGAUGUUCAUCGAAGACGAUGCGCAGUGAGUA-AAGACAUUUAUAAUAUAA ------------.....((.(((...(((.(((((....))))).))).))))).-.................. ( -10.90, z-score = -1.55, R) >droEre2.scaffold_4929 8562466 61 - 26641161 ------------AACAGGCAGCAAGAUGUUCAUCGAAGACGAUGCGCAGUGAGUA-AAGACUUUUAUUAUAUAA ------------.........((...(((.(((((....))))).))).))....-.................. ( -8.50, z-score = 0.32, R) >consensus ____________AAAAGGCAGCAAGAUGUUCAUCGAAGAUGAUGCGCAGUAAGUA_AACAUAUCUAUUAUA___ ..........................(((.(((((....))))).))).......................... ( -6.27 = -6.12 + -0.16)
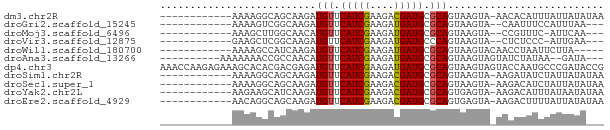


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:08:34 2011